An integrated, structure- and energy-based view of the genetic code
- PMID: 27448410
- PMCID: PMC5041475
- DOI: 10.1093/nar/gkw608
An integrated, structure- and energy-based view of the genetic code
Abstract
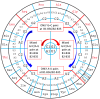
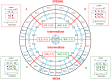
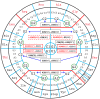
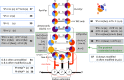
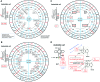
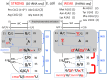
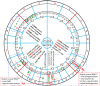
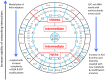
The principles of mRNA decoding are conserved among all extant life forms. We present an integrative view of all the interaction networks between mRNA, tRNA and rRNA: the intrinsic stability of codon-anticodon duplex, the conformation of the anticodon hairpin, the presence of modified nucleotides, the occurrence of non-Watson-Crick pairs in the codon-anticodon helix and the interactions with bases of rRNA at the A-site decoding site. We derive a more information-rich, alternative representation of the genetic code, that is circular with an unsymmetrical distribution of codons leading to a clear segregation between GC-rich 4-codon boxes and AU-rich 2:2-codon and 3:1-codon boxes. All tRNA sequence variations can be visualized, within an internal structural and energy framework, for each organism, and each anticodon of the sense codons. The multiplicity and complexity of nucleotide modifications at positions 34 and 37 of the anticodon loop segregate meaningfully, and correlate well with the necessity to stabilize AU-rich codon-anticodon pairs and to avoid miscoding in split codon boxes. The evolution and expansion of the genetic code is viewed as being originally based on GC content with progressive introduction of A/U together with tRNA modifications. The representation we present should help the engineering of the genetic code to include non-natural amino acids.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

