Gut microbiota-host interactions and juvenile idiopathic arthritis
- PMID: 27448997
- PMCID: PMC4957868
- DOI: 10.1186/s12969-016-0104-6
Gut microbiota-host interactions and juvenile idiopathic arthritis
Abstract
Background: Juvenile idiopathic arthritis is the most common form of chronic arthritis in children. There is mounting evidence that the microbiota may influence the disease.
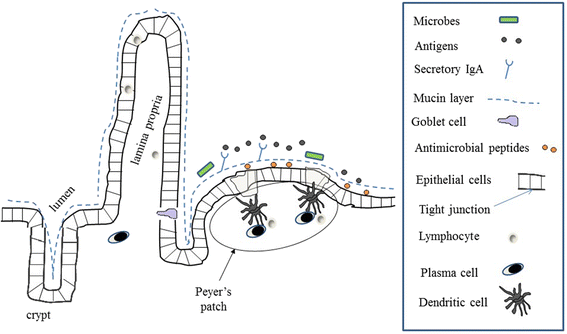
Main body: Recent observations in several systemic inflammatory diseases including JIA have indicated that abnormalities in the contents of the microbiota may be factors in disease pathogenesis, while other studies in turn have shown that environmental factors impacting the composition of the microbiota, such as delivery mode and early exposure to antibiotics, affect the risk of chronic inflammatory diseases including JIA. Microbial alterations may predispose to JIA through a variety of mechanisms, including impaired immunologic development, alterations in the balances of pro- versus anti-inflammatory bacteria, and low-grade mucosal inflammation. Additional confirmatory studies of microbiota aberrations and their risk factors are needed, as well as additional mechanistic studies linking these alterations to the disease itself.
Conclusions: The microbiota may influence the risk of JIA and other systemic inflammatory conditions through a variety of mechanisms. Additional research is required to improve our understanding of the links between the microbiota and arthritis, and the treatment implications thereof.
Keywords: Antibiotics; Juvenile arthritis; Microbiota.
Figures
References
-
- Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International league of associations for rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31(2):390–392. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

