Transcriptome-wide identification of mRNAs and lincRNAs associated with trastuzumab-resistance in HER2-positive breast cancer
- PMID: 27449296
- PMCID: PMC5288181
- DOI: 10.18632/oncotarget.10637
Transcriptome-wide identification of mRNAs and lincRNAs associated with trastuzumab-resistance in HER2-positive breast cancer
Abstract
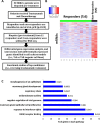
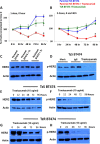
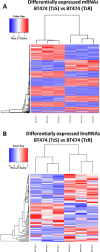
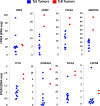
Approximately, 25-30% of early-stage breast tumors are classified at the molecular level as HER2-positive, which is an aggressive subtype of breast cancer. Amplification of the HER2 gene in these tumors results in a substantial increase in HER2 mRNA levels, and consequently, HER2 protein levels. HER2, a transmembrane receptor tyrosine kinase (RTK), is targeted therapeutically by a monoclonal antibody, trastuzumab (Tz), which has dramatically improved the prognosis of HER2-driven breast cancers. However, ~30% of patients develop resistance to trastuzumab and recur; and nearly all patients with advanced disease develop resistance over time and succumb to the disease. Mechanisms of trastuzumab resistance (TzR) are not well understood, although some studies suggest that growth factor signaling through other receptors may be responsible. However, these studies were based on cell culture models of the disease, and thus, it is not known which pathways are driving the resistance in vivo. Using an integrative transcriptomic approach of RNA isolated from trastuzumab-sensitive and trastuzumab-resistant HER2+ tumors, and isogenic cell culture models, we identified a small set of mRNAs and lincRNAs that are associated with trastuzumab-resistance (TzR). Functional analysis of a top candidate gene, S100P, demonstrated that inhibition of S100P results in reversing TzR. Mechanistically, S100P activates the RAS/MEK/MAPK pathway to compensate for HER2 inhibition by trastuzumab. Finally, we demonstrated that the upregulation of S100P appears to be driven by epigenomic changes at the enhancer level. Our current findings should pave the path toward new therapies for breast cancer patients.
Keywords: HER2; breast cancer; cancer therapy; drug resistance; trastuzumab-resistance.
Conflict of interest statement
All authors declare that there are no conflicts of interest to report.
Figures
Similar articles
-
Integrative transcriptome-wide analyses reveal critical HER2-regulated mRNAs and lincRNAs in HER2+ breast cancer.Breast Cancer Res Treat. 2015 Apr;150(2):321-34. doi: 10.1007/s10549-015-3327-1. Epub 2015 Mar 8. Breast Cancer Res Treat. 2015. PMID: 25749757
-
CTMP, a predictive biomarker for trastuzumab resistance in HER2-enriched breast cancer patient.Oncotarget. 2017 May 2;8(18):29699-29710. doi: 10.18632/oncotarget.10719. Oncotarget. 2017. PMID: 27447863 Free PMC article.
-
TNFα-Induced Mucin 4 Expression Elicits Trastuzumab Resistance in HER2-Positive Breast Cancer.Clin Cancer Res. 2017 Feb 1;23(3):636-648. doi: 10.1158/1078-0432.CCR-16-0970. Epub 2016 Oct 3. Clin Cancer Res. 2017. PMID: 27698002
-
Recent Insights into the Development of Preclinical Trastuzumab- Resistant HER2+ Breast Cancer Models.Curr Med Chem. 2018;25(17):1976-1998. doi: 10.2174/0929867323666161216144659. Curr Med Chem. 2018. PMID: 27993109 Review.
-
Recent advances in HER2 positive breast cancer epigenetics: Susceptibility and therapeutic strategies.Eur J Med Chem. 2017 Dec 15;142:316-327. doi: 10.1016/j.ejmech.2017.07.075. Epub 2017 Aug 1. Eur J Med Chem. 2017. PMID: 28800870 Review.
Cited by
-
Association between genome-wide epigenetic and genetic alterations in breast cancer tissue and response to HER2-targeted therapies in HER2-positive breast cancer patients: new findings and a systematic review.Cancer Drug Resist. 2022 Nov 2;5(4):995-1015. doi: 10.20517/cdr.2022.63. eCollection 2022. Cancer Drug Resist. 2022. PMID: 36627894 Free PMC article. Review.
-
YAP increases response to Trastuzumab in HER2-positive Breast Cancer by enhancing P73-induced apoptosis.J Cancer. 2020 Sep 25;11(22):6748-6759. doi: 10.7150/jca.48535. eCollection 2020. J Cancer. 2020. PMID: 33046997 Free PMC article.
-
A new mechanism of trastuzumab resistance in gastric cancer: MACC1 promotes the Warburg effect via activation of the PI3K/AKT signaling pathway.J Hematol Oncol. 2016 Aug 31;9(1):76. doi: 10.1186/s13045-016-0302-1. J Hematol Oncol. 2016. PMID: 27581375 Free PMC article.
-
Application of Microfluidic Systems for Breast Cancer Research.Micromachines (Basel). 2022 Jan 20;13(2):152. doi: 10.3390/mi13020152. Micromachines (Basel). 2022. PMID: 35208277 Free PMC article. Review.
-
Identification of key pathways and genes in response to trastuzumab treatment in breast cancer using bioinformatics analysis.Oncotarget. 2018 Mar 5;9(63):32149-32160. doi: 10.18632/oncotarget.24605. eCollection 2018 Aug 14. Oncotarget. 2018. PMID: 30181805 Free PMC article.
References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Merry CR, McMahon S, Thompson CL, Miskimen KL, Harris LN, Khalil AM. Integrative transcriptome-wide analyses reveal critical HER2-regulated mRNAs and lincRNAs in HER2+ breast cancer. Breast Cancer Res Treat. 2015;150:321–334. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

