Dual randomization of oligonucleotides to reduce the bias in ribosome-profiling libraries
- PMID: 27450428
- PMCID: PMC5024760
- DOI: 10.1016/j.ymeth.2016.07.011
Dual randomization of oligonucleotides to reduce the bias in ribosome-profiling libraries
Abstract
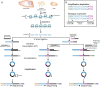
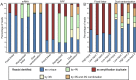
Protein translation is at the heart of cellular metabolism and its in-depth characterization is key for many lines of research. Recently, ribosome profiling became the state-of-the-art method to quantitatively characterize translation dynamics at a transcriptome-wide level. However, the strategy of library generation affects its outcomes. Here, we present a modified ribosome-profiling protocol starting from yeast, human cells and vertebrate brain tissue. We use a DNA linker carrying four randomized positions at its 5' end and a reverse-transcription (RT) primer with three randomized positions to reduce artifacts during library preparation. The use of seven randomized nucleotides allows to efficiently detect library-generation artifacts. We find that the effect of polymerase chain reaction (PCR) artifacts is relatively small for global analyses when sufficient input material is used. However, when input material is limiting, our strategy improves the sensitivity of gene-specific analyses. Furthermore, randomized nucleotides alleviate the skewed frequency of specific sequences at the 3' end of ribosome-protected fragments (RPFs) likely resulting from ligase specificity. Finally, strategies that rely on dual ligation show a high degree of gene-coverage variation. Taken together, our approach helps to remedy two of the main problems associated with ribosome-profiling data. This will facilitate the analysis of translational dynamics and increase our understanding of the influence of RNA modifications on translation.
Keywords: Codon-translation speed; RNA modification; Ribosome profiling; Sequencing bias; Translation; Translational control.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Transcriptome-wide measurement of translation by ribosome profiling.Methods. 2017 Aug 15;126:112-129. doi: 10.1016/j.ymeth.2017.05.028. Epub 2017 Jun 1. Methods. 2017. PMID: 28579404 Free PMC article.
-
Streamlined and sensitive mono- and di-ribosome profiling in yeast and human cells.Nat Methods. 2023 Nov;20(11):1704-1715. doi: 10.1038/s41592-023-02028-1. Epub 2023 Oct 2. Nat Methods. 2023. PMID: 37783882 Free PMC article.
-
Genome-wide translational profiling by ribosome footprinting.Methods Enzymol. 2010;470:119-42. doi: 10.1016/S0076-6879(10)70006-9. Epub 2010 Mar 1. Methods Enzymol. 2010. PMID: 20946809
-
Translation Analysis at the Genome Scale by Ribosome Profiling.Methods Mol Biol. 2016;1361:105-24. doi: 10.1007/978-1-4939-3079-1_7. Methods Mol Biol. 2016. PMID: 26483019 Review.
-
Ribosomal profiling adds new coding sequences to the proteome.Biochem Soc Trans. 2015 Dec;43(6):1271-6. doi: 10.1042/BST20150170. Biochem Soc Trans. 2015. PMID: 26614672 Review.
Cited by
-
RiboVIEW: a computational framework for visualization, quality control and statistical analysis of ribosome profiling data.Nucleic Acids Res. 2020 Jan 24;48(2):e7. doi: 10.1093/nar/gkz1074. Nucleic Acids Res. 2020. PMID: 31777932 Free PMC article.
-
XPRESSyourself: Enhancing, standardizing, and automating ribosome profiling computational analyses yields improved insight into data.PLoS Comput Biol. 2020 Jan 31;16(1):e1007625. doi: 10.1371/journal.pcbi.1007625. eCollection 2020 Jan. PLoS Comput Biol. 2020. PMID: 32004313 Free PMC article.
-
Principles, challenges, and advances in ribosome profiling: from bulk to low-input and single-cell analysis.Adv Biotechnol (Singap). 2023 Dec 1;1(4):6. doi: 10.1007/s44307-023-00006-4. Adv Biotechnol (Singap). 2023. PMID: 39883220 Free PMC article. Review.
-
Transcriptome-wide measurement of translation by ribosome profiling.Methods. 2017 Aug 15;126:112-129. doi: 10.1016/j.ymeth.2017.05.028. Epub 2017 Jun 1. Methods. 2017. PMID: 28579404 Free PMC article.
-
Nano LC-MS using capillary columns enables accurate quantification of modified ribonucleosides at low femtomol levels.RNA. 2018 Oct;24(10):1403-1417. doi: 10.1261/rna.065482.117. Epub 2018 Jul 16. RNA. 2018. PMID: 30012570 Free PMC article.
References
-
- Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In: Grosjean H., editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 2009. pp. 1–18.
-
- Agris P.F., Vendeix F.A.P., Graham W.D. TRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

