Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia
- PMID: 27450777
- PMCID: PMC5026943
- DOI: 10.1016/j.schres.2016.07.006
Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia
Abstract
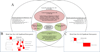
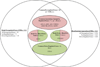
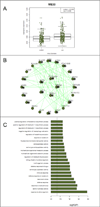
The application of microarray technology in schizophrenia research was heralded as paradigm-shifting, as it allowed for high-throughput assessment of cell and tissue function. This technology was widely adopted, initially in studies of postmortem brain tissue, and later in studies of peripheral blood. The collective body of schizophrenia microarray literature contains apparent inconsistencies between studies, with failures to replicate top hits, in part due to small sample sizes, cohort-specific effects, differences in array types, and other confounders. In an attempt to summarize existing studies of schizophrenia cases and non-related comparison subjects, we performed two mega-analyses of a combined set of microarray data from postmortem prefrontal cortices (n=315) and from ex-vivo blood tissues (n=578). We adjusted regression models per gene to remove non-significant covariates, providing best-estimates of transcripts dysregulated in schizophrenia. We also examined dysregulation of functionally related gene sets and gene co-expression modules, and assessed enrichment of cell types and genetic risk factors. The identities of the most significantly dysregulated genes were largely distinct for each tissue, but the findings indicated common emergent biological functions (e.g. immunity) and regulatory factors (e.g., predicted targets of transcription factors and miRNA species across tissues). Our network-based analyses converged upon similar patterns of heightened innate immune gene expression in both brain and blood in schizophrenia. We also constructed generalizable machine-learning classifiers using the blood-based microarray data. Our study provides an informative atlas for future pathophysiologic and biomarker studies of schizophrenia.
Keywords: Blood; Brain; Gene expression; Innate immunity; Random forests; Schizophrenia; Support vector machine; Transcriptome.
Published by Elsevier B.V.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

References
-
- Åberg K, Saetre P, Lindholm E, Ekholm B, Pettersson U, Adolfsson R, Jazin E. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am. J. Med. Genet. - Neuropsychiatr. Genet. 2006;141 B(1):84–90. - PubMed
-
- Arnedo J, Svrakic DM, Del Val C, Romero-Zaliz R, Hernendez-Cuervo H, Fanous AH, Pato MT, Pato CN, De Erausquin GA, Robert Cloninger C, Zwir I. Uncovering the hidden risk architecture of the schizophrenias: Confirmation in three independent genome-wide association studies. Am. J. Psychiatry. 2015;172(2):139–153. - PubMed
-
- Bergon A, Belzeaux R, Comte M, Pelletier F, Herve M, Gardiner EJ, Beveridge NJ, Liu B, Carr V, Scott RJ, Kelly B, Cairns MJ, Kumarasinghe N, Schall U, Blin O, Boucraut J, Tooney PA, Fakra E, Ibrahim EC. CX3CR1 is dysregulated in blood and brain from schizophrenia patients. Schizophr. Res. 2015;168(1–2):434–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

