The Microtubule Plus End Tracking Protein TIP150 Interacts with Cortactin to Steer Directional Cell Migration
- PMID: 27451391
- PMCID: PMC5034059
- DOI: 10.1074/jbc.M116.732719
The Microtubule Plus End Tracking Protein TIP150 Interacts with Cortactin to Steer Directional Cell Migration
Abstract
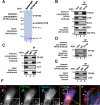
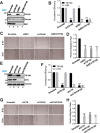
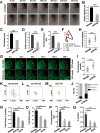
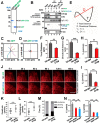
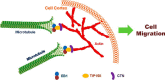
Cell migration is orchestrated by dynamic interactions of microtubules with the plasma membrane cortex. How these interactions facilitate these dynamic processes is still being actively investigated. TIP150 is a newly characterized microtubule plus end tracking protein essential for mitosis and entosis (Ward, T., Wang, M., Liu, X., Wang, Z., Xia, P., Chu, Y., Wang, X., Liu, L., Jiang, K., Yu, H., Yan, M., Wang, J., Hill, D. L., Huang, Y., Zhu, T., and Yao, X. (2013) Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288, 15771-15785; Xia, P., Zhou, J., Song, X., Wu, B., Liu, X., Li, D., Zhang, S., Wang, Z., Yu, H., Ward, T., Zhang, J., Li, Y., Wang, X., Chen, Y., Guo, Z., and Yao, X. (2014) Aurora A orchestrates entosis by regulating a dynamic MCAK-TIP150 interaction. J. Mol. Cell Biol. 6, 240-254). Here we show that TIP150 links dynamic microtubules to steer cell migration by interacting with cortactin. Mechanistically, TIP150 binds to cortactin via its C-terminal tail. Interestingly, the C-terminal TIP150 proline-rich region (CT150) binds to the Src homology 3 domain of cortactin specifically, and such an interaction is negatively regulated by EGF-elicited tyrosine phosphorylation of cortactin. Importantly, suppression of TIP150 or overexpression of phospho-mimicking cortactin inhibits polarized cell migration. In addition, CT150 disrupts the biochemical interaction between TIP150 and cortactin in vitro, and perturbation of the TIP150-cortactin interaction in vivo using a membrane-permeable TAT-CT150 peptide results in an inhibition of directional cell migration. We reason that a dynamic TIP150-cortactin interaction orchestrates directional cell migration via coupling dynamic microtubule plus ends to the cortical cytoskeleton.
Keywords: cell invasion; cell migration; cell motility; cellular regulation; microtubule; microtubule-associated protein (MAP).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Cosen-Binker L. I., and Kapus A. (2006) Cortactin: the gray eminence of the cytoskeleton. Physiology 21, 352–361 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

