Transient Tissue-Scale Deformation Coordinates Alignment of Planar Cell Polarity Junctions in the Mammalian Skin
- PMID: 27451904
- PMCID: PMC5005808
- DOI: 10.1016/j.cub.2016.06.030
Transient Tissue-Scale Deformation Coordinates Alignment of Planar Cell Polarity Junctions in the Mammalian Skin
Abstract
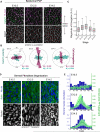
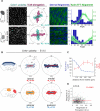
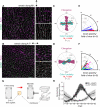
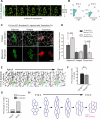
Planar cell polarity (PCP) refers to the collective alignment of polarity along the tissue plane. In skin, the largest mammalian organ, PCP aligns over extremely long distances, but the global cues that orient tissue polarity are unknown. Here, we show that Celsr1 asymmetry arises concomitant with a gradient of tissue deformation oriented along the medial-lateral axis. This uniaxial tissue tension, whose origin remains unknown, transiently transforms basal epithelial cells from initially isotropic and disordered states into highly elongated and aligned morphologies. Reorienting tissue deformation is sufficient to shift the global axis of polarity, suggesting that uniaxial tissue strain can act as a long-range polarizing cue. Observations both in vivo and in vitro suggest that the effect of tissue anisotropy on Celsr1 polarity is not a direct consequence of cell shape but rather reflects the restructuring of cell-cell interfaces during oriented cell divisions and cell rearrangements that serve to relax tissue strain. We demonstrate that cell intercalations remodel intercellular junctions predominantly between the mediolateral interfaces of neighboring cells. This restructuring of the cell surface polarizes Celsr1, which is slow to accumulate at nascent junctions yet stably associates with persistent junctions. We propose that tissue anisotropy globally aligns Celsr1 polarity by creating a directional bias in the formation of new cell interfaces while simultaneously aligning the persistent interfaces at which Celsr1 prefers to accumulate.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

References
-
- Shimada Y, Usui T, Yanagawa S, Takeichi M, Uemura T. Asymmetric colocalization of Flamingo, a seven-pass transmembrane cadherin, and Dishevelled in planar cell polarization. Curr Biol. 2001;11:859–863. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

