PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kα Inhibition
- PMID: 27451907
- PMCID: PMC4982440
- DOI: 10.1016/j.ccell.2016.06.004
PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kα Inhibition
Abstract
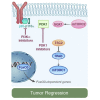
PIK3CA, which encodes the p110α subunit of PI3K, is frequently mutated and oncogenic in breast cancer. PI3Kα inhibitors are in clinical development and despite promising early clinical activity, intrinsic resistance is frequent among patients. We have previously reported that residual downstream mTORC1 activity upon treatment with PI3Kα inhibitors drives resistance to these agents. However, the mechanism underlying this phenotype is not fully understood. Here we show that in cancer cells resistant to PI3Kα inhibition, PDK1 blockade restores sensitivity to these therapies. SGK1, which is activated by PDK1, contributes to the maintenance of residual mTORC1 activity through direct phosphorylation and inhibition of TSC2. Targeting either PDK1 or SGK1 prevents mTORC1 activation, restoring the antitumoral effects of PI3Kα inhibition in resistant cells.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Alessi D.R., James S.R., Downes C.P., Holmes A.B., Gaffney P.R., Reese C.B., Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997;7:261–269. - PubMed
-
- Alessi D.R., Pearce L.R., Garcia-Martinez J.M. New insights into mTOR signaling: mTORC2 and beyond. Sci. Signal. 2009;2:pe27. - PubMed
-
- Arencibia J.M., Pastor-Flores D., Bauer A.F., Schulze J.O., Biondi R.M. AGC protein kinases: from structural mechanism of regulation to allosteric drug development for the treatment of human diseases. Biochim. Biophys. Acta. 2013;1834:1302–1321. - PubMed
-
- Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

