Inhibition of the Polyamine Synthesis Pathway Is Synthetically Lethal with Loss of Argininosuccinate Synthase 1
- PMID: 27452468
- PMCID: PMC4978703
- DOI: 10.1016/j.celrep.2016.06.097
Inhibition of the Polyamine Synthesis Pathway Is Synthetically Lethal with Loss of Argininosuccinate Synthase 1
Abstract
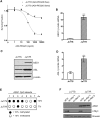
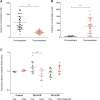
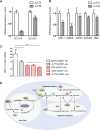
Argininosuccinate synthase 1 (ASS1) is the rate-limiting enzyme for arginine biosynthesis. ASS1 expression is lost in a range of tumor types, including 50% of malignant pleural mesotheliomas. Starving ASS1-deficient cells of arginine with arginine blockers such as ADI-PEG20 can induce selective lethality and has shown great promise in the clinical setting. We have generated a model of ADI-PEG20 resistance in mesothelioma cells. This resistance is mediated through re-expression of ASS1 via demethylation of the ASS1 promoter. Through coordinated transcriptomic and metabolomic profiling, we have shown that ASS1-deficient cells have decreased levels of acetylated polyamine metabolites, together with a compensatory increase in the expression of polyamine biosynthetic enzymes. Upon arginine deprivation, polyamine metabolites are decreased in the ASS1-deficient cells and in plasma isolated from ASS1-deficient mesothelioma patients. We identify a synthetic lethal dependence between ASS1 deficiency and polyamine metabolism, which could potentially be exploited for the treatment of ASS1-negative cancers.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Allen M.D., Luong P., Hudson C., Leyton J., Delage B., Ghazaly E., Cutts R., Yuan M., Syed N., Lo Nigro C. Prognostic and therapeutic impact of argininosuccinate synthetase 1 control in bladder cancer as monitored longitudinally by PET imaging. Cancer Res. 2014;74:896–907. - PubMed
-
- Bachrach U., Wang Y.C., Tabib A. Polyamines: new cues in cellular signal transduction. News Physiol. Sci. 2001;16:106–109. - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B. 1995;57:289–300.
-
- Brabender J., Lord R.V., Danenberg K.D., Metzger R., Schneider P.M., Uetake H., Kawakami K., Park J.M., Salonga D., Peters J.H. Upregulation of ornithine decarboxylase mRNA expression in Barrett’s esophagus and Barrett’s-associated adenocarcinoma. J. Gastrointest. Surg. 2001;5:174–181, discussion 182. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

