A high-content image-based method for quantitatively studying context-dependent cell population dynamics
- PMID: 27452732
- PMCID: PMC4958988
- DOI: 10.1038/srep29752
A high-content image-based method for quantitatively studying context-dependent cell population dynamics
Abstract
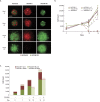
Tumor progression results from a complex interplay between cellular heterogeneity, treatment response, microenvironment and heterocellular interactions. Existing approaches to characterize this interplay suffer from an inability to distinguish between multiple cell types, often lack environmental context, and are unable to perform multiplex phenotypic profiling of cell populations. Here we present a high-throughput platform for characterizing, with single-cell resolution, the dynamic phenotypic responses (i.e. morphology changes, proliferation, apoptosis) of heterogeneous cell populations both during standard growth and in response to multiple, co-occurring selective pressures. The speed of this platform enables a thorough investigation of the impacts of diverse selective pressures including genetic alterations, therapeutic interventions, heterocellular components and microenvironmental factors. The platform has been applied to both 2D and 3D culture systems and readily distinguishes between (1) cytotoxic versus cytostatic cellular responses; and (2) changes in morphological features over time and in response to perturbation. These important features can directly influence tumor evolution and clinical outcome. Our image-based approach provides a deeper insight into the cellular dynamics and heterogeneity of tumors (or other complex systems), with reduced reagents and time, offering advantages over traditional biological assays.
Figures




References
-
- Junttila M. R. & de Sauvage F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

