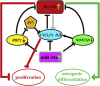
MiR-34a Promotes Osteogenic Differentiation of Human Adipose-Derived Stem Cells via the RBP2/NOTCH1/CYCLIN D1 Coregulatory Network
- PMID: 27453008
- PMCID: PMC4982986
- DOI: 10.1016/j.stemcr.2016.06.010
MiR-34a Promotes Osteogenic Differentiation of Human Adipose-Derived Stem Cells via the RBP2/NOTCH1/CYCLIN D1 Coregulatory Network
Abstract
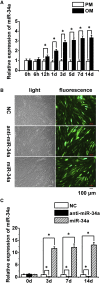
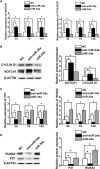
MiR-34a was demonstrated to be upregulated during the osteogenic differentiation of human adipose-derived stem cells (hASCs). Overexpression of miR-34a significantly increased alkaline phosphatase activity, mineralization capacity, and the expression of osteogenesis-associated genes in hASCs in vitro. Enhanced heterotopic bone formation in vivo was also observed upon overexpression of miR-34a in hASCs. Mechanistic investigations revealed that miR-34a inhibited the expression of retinoblastoma binding protein 2 (RBP2) and reduced the luciferase activity of reporter gene construct comprising putative miR-34a binding sites in the 3' UTR of RBP2. Moreover, miR-34a downregulated the expression of NOTCH1 and CYCLIN D1 and upregulated the expression of RUNX2 by targeting RBP2, NOTCH1, and CYCLIN D1. Taken together, our results suggested that miR-34a promotes the osteogenic differentiation of hASCs via the RBP2/NOTCH1/CYCLIN D1 coregulatory network, indicating that miR-34a-targeted therapy could be a valuable approach to promote bone regeneration.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bosnakovski D., Mizuno M., Kim G., Takagi S., Okumura M., Fujinaga T. Isolation and multilineage differentiation of bovine bone marrow mesenchymal stem cells. Cell Tissue Res. 2005;319:243–253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

