CRISPR-Barcoding for Intratumor Genetic Heterogeneity Modeling and Functional Analysis of Oncogenic Driver Mutations
- PMID: 27453044
- PMCID: PMC5537739
- DOI: 10.1016/j.molcel.2016.06.017
CRISPR-Barcoding for Intratumor Genetic Heterogeneity Modeling and Functional Analysis of Oncogenic Driver Mutations
Abstract
Intratumor genetic heterogeneity underlies the ability of tumors to evolve and adapt to different environmental conditions. Using CRISPR/Cas9 technology and specific DNA barcodes, we devised a strategy to recapitulate and trace the emergence of subpopulations of cancer cells containing a mutation of interest. We used this approach to model different mechanisms of lung cancer cell resistance to EGFR inhibitors and to assess effects of combined drug therapies. By overcoming intrinsic limitations of current approaches, CRISPR-barcoding also enables investigation of most types of genetic modifications, including repair of oncogenic driver mutations. Finally, we used highly complex barcodes inserted at a specific genome location as a means of simultaneously tracing the fates of many thousands of genetically labeled cancer cells. CRISPR-barcoding is a straightforward and highly flexible method that should greatly facilitate the functional investigation of specific mutations, in a context that closely mimics the complexity of cancer.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


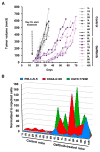
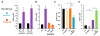

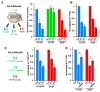
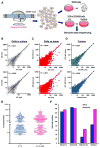
Comment in
-
Who Is in the Driver's Seat: Tracing Cancer Genes Using CRISPR-Barcoding.Mol Cell. 2016 Aug 4;63(3):352-4. doi: 10.1016/j.molcel.2016.07.013. Mol Cell. 2016. PMID: 27494556
-
CRISPR-barcoding in non small cell lung cancer: from intratumor genetic heterogeneity modeling to cancer therapy application.J Thorac Dis. 2017 Jul;9(7):1759-1762. doi: 10.21037/jtd.2017.06.27. J Thorac Dis. 2017. PMID: 28839956 Free PMC article. No abstract available.
References
-
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. - PubMed
-
- Bhang HE, Ruddy DA, Krishnamurthy Radhakrishna V, Caushi JX, Zhao R, Hims MM, Singh AP, Kao I, Rakiec D, Shaw P, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med. 2015;21:440–448. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

