Comprehensive Identification of RNA-Binding Domains in Human Cells
- PMID: 27453046
- PMCID: PMC5003815
- DOI: 10.1016/j.molcel.2016.06.029
Comprehensive Identification of RNA-Binding Domains in Human Cells
Abstract
Mammalian cells harbor more than a thousand RNA-binding proteins (RBPs), with half of these employing unknown modes of RNA binding. We developed RBDmap to determine the RNA-binding sites of native RBPs on a proteome-wide scale. We identified 1,174 binding sites within 529 HeLa cell RBPs, discovering numerous RNA-binding domains (RBDs). Catalytic centers or protein-protein interaction domains are in close relationship with RNA-binding sites, invoking possible effector roles of RNA in the control of protein function. Nearly half of the RNA-binding sites map to intrinsically disordered regions, uncovering unstructured domains as prevalent partners in protein-RNA interactions. RNA-binding sites represent hot spots for defined posttranslational modifications such as lysine acetylation and tyrosine phosphorylation, suggesting metabolic and signal-dependent regulation of RBP function. RBDs display a high degree of evolutionary conservation and incidence of Mendelian mutations, suggestive of important functional roles. RBDmap thus yields profound insights into native protein-RNA interactions in living cells.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
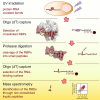


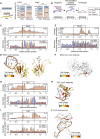



Comment in
-
From Protein-RNA Predictions toward a Peptide-RNA Code.Mol Cell. 2016 Nov 3;64(3):437-438. doi: 10.1016/j.molcel.2016.10.023. Mol Cell. 2016. PMID: 27814488
References
-
- Baltz A.G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. - PubMed
-
- Barbalat R., Ewald S.E., Mouchess M.L., Barton G.M. Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 2011;29:185–214. - PubMed
-
- Boersema P.J., Aye T.T., van Veen T.A., Heck A.J., Mohammed S. Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics. 2008;8:4624–4632. - PubMed
-
- Bono F., Ebert J., Lorentzen E., Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

