Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks
- PMID: 27453047
- PMCID: PMC4992412
- DOI: 10.1016/j.molcel.2016.06.020
Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks
Abstract
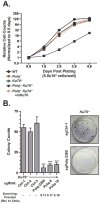
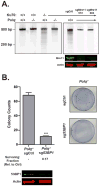
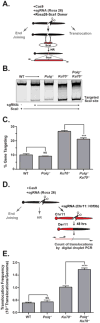
DNA polymerase theta (Pol θ)-mediated end joining (TMEJ) has been implicated in the repair of chromosome breaks, but its cellular mechanism and role relative to canonical repair pathways are poorly understood. We show that it accounts for most repairs associated with microhomologies and is made efficient by coupling a microhomology search to removal of non-homologous tails and microhomology-primed synthesis across broken ends. In contrast to non-homologous end joining (NHEJ), TMEJ efficiently repairs end structures expected after aborted homology-directed repair (5' to 3' resected ends) or replication fork collapse. It typically does not compete with canonical repair pathways but, in NHEJ-deficient cells, is engaged more frequently and protects against translocation. Cell viability is also severely impaired upon combined deficiency in Pol θ and a factor that antagonizes end resection (Ku or 53BP1). TMEJ thus helps to sustain cell viability and genome stability by rescuing chromosome break repair when resection is misregulated or NHEJ is compromised.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
DNA Polymerase θ: Duct Tape and Zip Ties for a Fragile Genome.Mol Cell. 2016 Aug 18;63(4):542-544. doi: 10.1016/j.molcel.2016.08.006. Mol Cell. 2016. PMID: 27540853
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

