Mutations in Citron Kinase Cause Recessive Microlissencephaly with Multinucleated Neurons
- PMID: 27453579
- PMCID: PMC4974106
- DOI: 10.1016/j.ajhg.2016.07.003
Mutations in Citron Kinase Cause Recessive Microlissencephaly with Multinucleated Neurons
Abstract
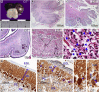
Primary microcephaly is a neurodevelopmental disorder that is caused by a reduction in brain size as a result of defects in the proliferation of neural progenitor cells during development. Mutations in genes encoding proteins that localize to the mitotic spindle and centrosomes have been implicated in the pathogenicity of primary microcephaly. In contrast, the contractile ring and midbody required for cytokinesis, the final stage of mitosis, have not previously been implicated by human genetics in the molecular mechanisms of this phenotype. Citron kinase (CIT) is a multi-domain protein that localizes to the cleavage furrow and midbody of mitotic cells, where it is required for the completion of cytokinesis. Rodent models of Cit deficiency highlighted the role of this gene in neurogenesis and microcephaly over a decade ago. Here, we identify recessively inherited pathogenic variants in CIT as the genetic basis of severe microcephaly and neonatal death. We present postmortem data showing that CIT is critical to building a normally sized human brain. Consistent with cytokinesis defects attributed to CIT, multinucleated neurons were observed throughout the cerebral cortex and cerebellum of an affected proband, expanding our understanding of mechanisms attributed to primary microcephaly.
Keywords: autosomal recessive; citron kinase; cytokinesis; lissencephaly; neurogenesis; primary microcephaly; splicing mutation.
Copyright © 2016 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bond J., Roberts E., Mochida G.H., Hampshire D.J., Scott S., Askham J.M., Springell K., Mahadevan M., Crow Y.J., Markham A.F. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 2002;32:316–320. - PubMed
-
- Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

