Real-time selective sequencing using nanopore technology
- PMID: 27454285
- PMCID: PMC5008457
- DOI: 10.1038/nmeth.3930
Real-time selective sequencing using nanopore technology
Abstract
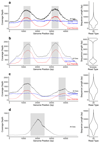
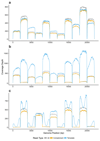
The Oxford Nanopore Technologies MinION sequencer enables the selection of specific DNA molecules for sequencing by reversing the driving voltage across individual nanopores. To directly select molecules for sequencing, we used dynamic time warping to match reads to reference sequences. We demonstrate our open-source Read Until software in real-time selective sequencing of regions within small genomes, individual amplicon enrichment and normalization of an amplicon set.
Conflict of interest statement
ML is a member of the MinION access program (MAP) and has received free-of-charge flow cells and kits for nanopore sequencing and travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences.
Figures

References
-
- Heron A, et al. METHOD FOR ATTACHING ONE OR MORE POLYNUCLEOTIDE BINDING PROTEINS TO A TARGET POLYNUCLEOTIDE. WO patent WO 2015/110813 A1. 2015
-
- Heron A, et al. MODIFIED HELICASES. US patent US 2015/0191709 A1. 2015
-
- Moysey R, Heron Andrew J. METHOD OF CHARACTERIZING A TARGET POLYNUCLEOTIDE USING A PORE AND A HEL308 HELICASE. WO patent WO 2013/057495 A3. 2013
-
- Urban JM, Bliss J, Lawrence CE, Gerbi SA. Sequencing ultra-long DNA molecules with the Oxford Nanopore MinION. bioRxiv. 2015 doi: 10.1101/019281. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

