E2f8 mediates tumor suppression in postnatal liver development
- PMID: 27454291
- PMCID: PMC4966321
- DOI: 10.1172/JCI85506
E2f8 mediates tumor suppression in postnatal liver development
Abstract
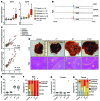
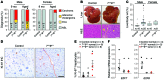
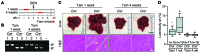
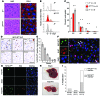
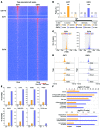
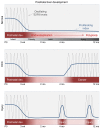
E2F-mediated transcriptional repression of cell cycle-dependent gene expression is critical for the control of cellular proliferation, survival, and development. E2F signaling also interacts with transcriptional programs that are downstream of genetic predictors for cancer development, including hepatocellular carcinoma (HCC). Here, we evaluated the function of the atypical repressor genes E2f7 and E2f8 in adult liver physiology. Using several loss-of-function alleles in mice, we determined that combined deletion of E2f7 and E2f8 in hepatocytes leads to HCC. Temporal-specific ablation strategies revealed that E2f8's tumor suppressor role is critical during the first 2 weeks of life, which correspond to a highly proliferative stage of postnatal liver development. Disruption of E2F8's DNA binding activity phenocopied the effects of an E2f8 null allele and led to HCC. Finally, a profile of chromatin occupancy and gene expression in young and tumor-bearing mice identified a set of shared targets for E2F7 and E2F8 whose increased expression during early postnatal liver development is associated with HCC progression in mice. Increased expression of E2F8-specific target genes was also observed in human liver biopsies from HCC patients compared to healthy patients. In summary, these studies suggest that E2F8-mediated transcriptional repression is a critical tumor suppressor mechanism during postnatal liver development.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

