Detection of 2-hydroxyglutarate in brain tumors by triple-refocusing MR spectroscopy at 3T in vivo
- PMID: 27454352
- PMCID: PMC5266748
- DOI: 10.1002/mrm.26347
Detection of 2-hydroxyglutarate in brain tumors by triple-refocusing MR spectroscopy at 3T in vivo
Abstract
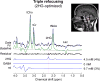
Purpose: To test the efficacy of triple-refocusing MR spectroscopy (MRS) for improved detection of 2-hydroxyglutarate (2HG) in brain tumors at 3T in vivo.
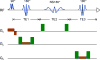
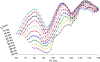
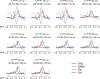
Methods: The triple-refocusing sequence parameters were tailored at 3T, with density-matrix simulations and phantom validation, for enhancing the 2HG 2.25-ppm signal selectivity with respect to the adjacent resonances of glutamate (Glu), glutamine (Gln), and gamma-aminobutyric acid (GABA). In vivo MRS data were acquired from 15 glioma patients and analyzed with LCModel using calculated basis spectra. Metabolites were quantified with reference to water.
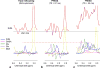
Results: A triple-refocusing sequence (echo time = 137 ms) was obtained for 2HG detection. The 2HG 2.25-ppm signal was large and narrow while the Glu and Gln signals between 2.2 and 2.3 ppm were minimal. The optimized triple refocusing offered improved separation of 2HG from Glu, Gln and GABA when compared with published MRS methods. 2HG was detected in all 15 patients, the estimated 2HG concentrations ranging from 2.4 to 15.0 mM, with Cramer-Rao lower bounds of 2%-11%. The 2HG estimates did not show significant correlation with total choline.
Conclusion: The optimized triple refocusing provides excellent 2HG signal discrimination from adjacent resonances and may confer reliable in vivo measurement of 2HG at relatively low concentrations. Magn Reson Med 78:40-48, 2017. © 2016 International Society for Magnetic Resonance in Medicine.
Keywords: 1H MRS; 2-hydroxyglutarate; 3T; glioma; human brain tumor; isocitrate dehydrogenase; triple refocusing.
© 2016 International Society for Magnetic Resonance in Medicine.
Figures





References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. - PubMed
-
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

