The GlnR Regulon in Streptococcus mutans Is Differentially Regulated by GlnR and PmrA
- PMID: 27454482
- PMCID: PMC4959772
- DOI: 10.1371/journal.pone.0159599
The GlnR Regulon in Streptococcus mutans Is Differentially Regulated by GlnR and PmrA
Abstract
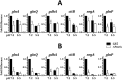
GlnR-mediated repression of the GlnR regulon at acidic pH is required for optimal acid tolerance in Streptococcus mutans, the etiologic agent for dental caries. Unlike most streptococci, the GlnR regulon is also regulated by newly identified PmrA (SMUGS5_RS05810) at the transcriptional level in S. mutans GS5. Results from gel mobility shift assays confirmed that both GlnR and PmrA recognized the putative GlnR box in the promoter regions of the GlnR regulon genes. By using a chemostat culture system, we found that PmrA activated the expression of the GlnR regulon at pH 7, and that this activation was enhanced by excess glucose. Deletion of pmrA (strain ΔPmrA) reduced the survival rate of S. mutans GS5 at pH 3 moderately, whereas the GlnR mutant (strain ΔGlnR) exhibited an acid-sensitive phenotype in the acid killing experiments. Elevated biofilm formation in both ΔGlnR and ΔPmrA mutant strains is likely a result of indirect regulation of the GlnR regulon since GlnR and PmrA regulate the regulon differently. Taken together, it is suggested that activation of the GlnR regulon by PmrA at pH 7 ensures adequate biosynthesis of amino acid precursor, whereas repression by GlnR at acidic pH allows greater ATP generation for acid tolerance. The tight regulation of the GlnR regulon in response to pH provides an advantage for S. mutans to better survive in its primary niche, the oral cavity.
Conflict of interest statement
Figures



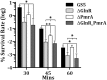


References
-
- Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;7(1):95–107. Epub 2004/12/08. . - PubMed
-
- Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 2000;193(1):1–6. . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

