Protein-protein interaction extraction with feature selection by evaluating contribution levels of groups consisting of related features
- PMID: 27454611
- PMCID: PMC4965725
- DOI: 10.1186/s12859-016-1100-z
Protein-protein interaction extraction with feature selection by evaluating contribution levels of groups consisting of related features
Abstract
Background: Protein-protein interaction (PPI) extraction from published scientific articles is one key issue in biological research due to its importance in grasping biological processes. Despite considerable advances of recent research in automatic PPI extraction from articles, demand remains to enhance the performance of the existing methods.
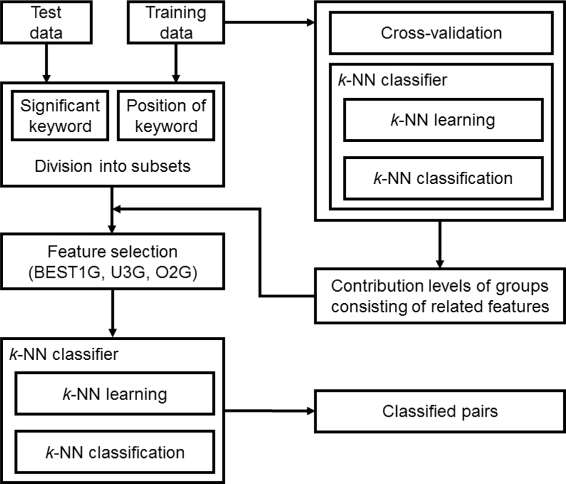
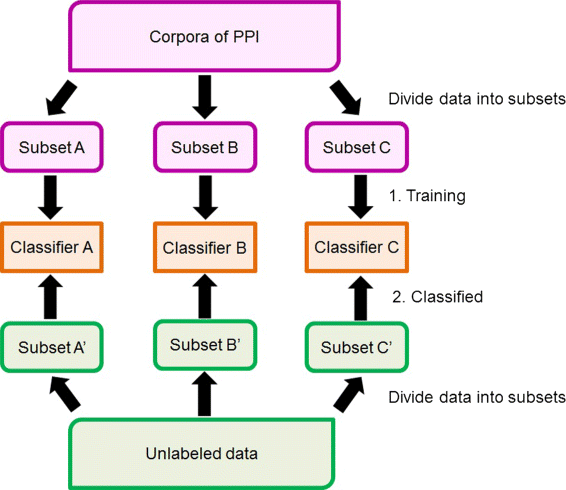
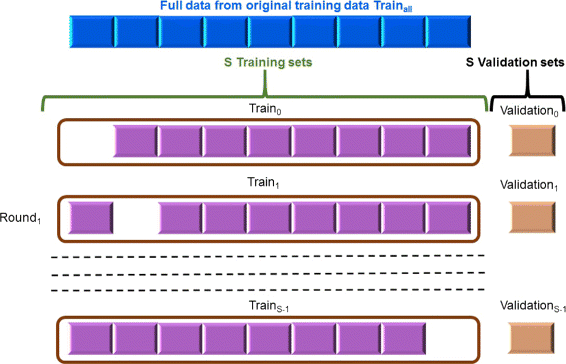
Results: Our feature-based method incorporates the strength of many kinds of diverse features, such as lexical and word context features derived from sentences, syntactic features derived from parse trees, and features using existing patterns to extract PPIs automatically from articles. Among these abundant features, we assemble the related features into four groups and define the contribution level (CL) for each group, which consists of related features. Our method consists of two steps. First, we divide the training set into subsets based on the structure of the sentence and the existence of significant keywords (SKs) and apply the sentence patterns given in advance to each subset. Second, we automatically perform feature selection based on the CL values of the four groups that consist of related features and the k-nearest neighbor algorithm (k-NN) through three approaches: (1) focusing on the group with the best contribution level (BEST1G); (2) unoptimized combination of three groups with the best contribution levels (U3G); (3) optimized combination of two groups with the best contribution levels (O2G).
Conclusions: Our method outperforms other state-of-the-art PPI extraction systems in terms of F-score on the HPRD50 corpus and achieves promising results that are comparable with these PPI extraction systems on other corpora. Further, our method always obtains the best F-score on all the corpora than when using k-NN only without exploiting the CLs of the groups of related features.
Keywords: Biomedical text mining; Information extraction; Protein protein interaction; k-nearest neighbors.
Figures
Similar articles
-
Distributed smoothed tree kernel for protein-protein interaction extraction from the biomedical literature.PLoS One. 2017 Nov 3;12(11):e0187379. doi: 10.1371/journal.pone.0187379. eCollection 2017. PLoS One. 2017. PMID: 29099838 Free PMC article.
-
A hybrid approach to extract protein-protein interactions.Bioinformatics. 2011 Jan 15;27(2):259-65. doi: 10.1093/bioinformatics/btq620. Epub 2010 Nov 8. Bioinformatics. 2011. PMID: 21062765
-
Exploiting graph kernels for high performance biomedical relation extraction.J Biomed Semantics. 2018 Jan 30;9(1):7. doi: 10.1186/s13326-017-0168-3. J Biomed Semantics. 2018. PMID: 29382397 Free PMC article.
-
Discovering novel protein-protein interactions by measuring the protein semantic similarity from the biomedical literature.J Bioinform Comput Biol. 2014 Dec;12(6):1442008. doi: 10.1142/S0219720014420086. J Bioinform Comput Biol. 2014. PMID: 25385082
-
A comprehensive benchmark of kernel methods to extract protein-protein interactions from literature.PLoS Comput Biol. 2010 Jul 1;6(7):e1000837. doi: 10.1371/journal.pcbi.1000837. PLoS Comput Biol. 2010. PMID: 20617200 Free PMC article.
Cited by
-
HMNPPID-human malignant neoplasm protein-protein interaction database.Hum Genomics. 2019 Oct 22;13(Suppl 1):44. doi: 10.1186/s40246-019-0223-5. Hum Genomics. 2019. PMID: 31639057 Free PMC article.
-
Using a Large Margin Context-Aware Convolutional Neural Network to Automatically Extract Disease-Disease Association from Literature: Comparative Analytic Study.JMIR Med Inform. 2019 Nov 26;7(4):e14502. doi: 10.2196/14502. JMIR Med Inform. 2019. PMID: 31769759 Free PMC article.
-
Distributed smoothed tree kernel for protein-protein interaction extraction from the biomedical literature.PLoS One. 2017 Nov 3;12(11):e0187379. doi: 10.1371/journal.pone.0187379. eCollection 2017. PLoS One. 2017. PMID: 29099838 Free PMC article.
-
Leveraging prior knowledge for protein-protein interaction extraction with memory network.Database (Oxford). 2018 Jan 1;2018:bay071. doi: 10.1093/database/bay071. Database (Oxford). 2018. PMID: 30010731 Free PMC article.
-
Identification of conclusive association entities in biomedical articles.J Biomed Semantics. 2019 Jan 7;10(1):1. doi: 10.1186/s13326-018-0194-9. J Biomed Semantics. 2019. PMID: 30616688 Free PMC article.
References
-
- Liu B, Qian L, Wang H, Zhou G. Dependency-driven feature-based learning for extracting protein-protein interactions from biomedical text. In: Proceedings of the 23rd International Conference on Computational Linguistics (Coling 2010). Beijing, China: 2010. p. 757–765.
-
- Landeghem S, Saeys Y, Peer Y. Extracting protein-protein interactions from text using rich feature vectors and feature selection. In: Proceedings of the Third International Symposium on Semantic Mining in Biomedicine. Turku, Finland: 2008. p. 77–84.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

