A thrifty variant in CREBRF strongly influences body mass index in Samoans
- PMID: 27455349
- PMCID: PMC5069069
- DOI: 10.1038/ng.3620
A thrifty variant in CREBRF strongly influences body mass index in Samoans
Abstract
Samoans are a unique founder population with a high prevalence of obesity, making them well suited for identifying new genetic contributors to obesity. We conducted a genome-wide association study (GWAS) in 3,072 Samoans, discovered a variant, rs12513649, strongly associated with body mass index (BMI) (P = 5.3 × 10(-14)), and replicated the association in 2,102 additional Samoans (P = 1.2 × 10(-9)). Targeted sequencing identified a strongly associated missense variant, rs373863828 (p.Arg457Gln), in CREBRF (meta P = 1.4 × 10(-20)). Although this variant is extremely rare in other populations, it is common in Samoans (frequency of 0.259), with an effect size much larger than that of any other known common BMI risk variant (1.36-1.45 kg/m(2) per copy of the risk-associated allele). In comparison to wild-type CREBRF, the Arg457Gln variant when overexpressed selectively decreased energy use and increased fat storage in an adipocyte cell model. These data, in combination with evidence of positive selection of the allele encoding p.Arg457Gln, support a 'thrifty' variant hypothesis as a factor in human obesity.
Figures

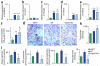
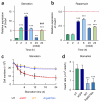
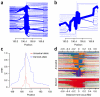
Comment in
-
Obesity: BMI-increasing variant identified in Samoans.Nat Rev Endocrinol. 2016 Oct;12(10):560. doi: 10.1038/nrendo.2016.129. Epub 2016 Aug 5. Nat Rev Endocrinol. 2016. PMID: 27494387 No abstract available.
-
CREBRF variant increases obesity risk and protects against diabetes in Samoans.Nat Genet. 2016 Aug 30;48(9):976-8. doi: 10.1038/ng.3653. Nat Genet. 2016. PMID: 27573685
References
-
- McGarvey ST. Obesity in Samoans and a perspective on its etiology in Polynesians. Am. J. Clin. Nutr. 1991;53(Suppl. 6):1586S–1594S. - PubMed
-
- McGarvey ST, Bindon JR, Crews DE, Schendel DE. In: Human Population Biology: A Transdisciplinary Science. Little MA, Haas JD, editors. Academic Press; 1989. pp. 263–279.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

