DNA polymerase ι functions in the generation of tandem mutations during somatic hypermutation of antibody genes
- PMID: 27455952
- PMCID: PMC4995076
- DOI: 10.1084/jem.20151227
DNA polymerase ι functions in the generation of tandem mutations during somatic hypermutation of antibody genes
Abstract
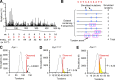
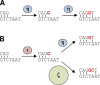
DNA polymerase ι (Pol ι) is an attractive candidate for somatic hypermutation in antibody genes because of its low fidelity. To identify a role for Pol ι, we analyzed mutations in two strains of mice with deficiencies in the enzyme: 129 mice with negligible expression of truncated Pol ι, and knock-in mice that express full-length Pol ι that is catalytically inactive. Both strains had normal frequencies and spectra of mutations in the variable region, indicating that loss of Pol ι did not change overall mutagenesis. We next examined if Pol ι affected tandem mutations generated by another error-prone polymerase, Pol ζ. The frequency of contiguous mutations was analyzed using a novel computational model to determine if they occur during a single DNA transaction or during two independent events. Analyses of 2,000 mutations from both strains indicated that Pol ι-compromised mice lost the tandem signature, whereas C57BL/6 mice accumulated significant amounts of double mutations. The results support a model where Pol ι occasionally accesses the replication fork to generate a first mutation, and Pol ζ extends the mismatch with a second mutation.
@2016.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

