Unravelling population genetic structure with mitochondrial DNA in a notional panmictic coastal crab species: sample size makes the difference
- PMID: 27455997
- PMCID: PMC4960869
- DOI: 10.1186/s12862-016-0720-2
Unravelling population genetic structure with mitochondrial DNA in a notional panmictic coastal crab species: sample size makes the difference
Abstract
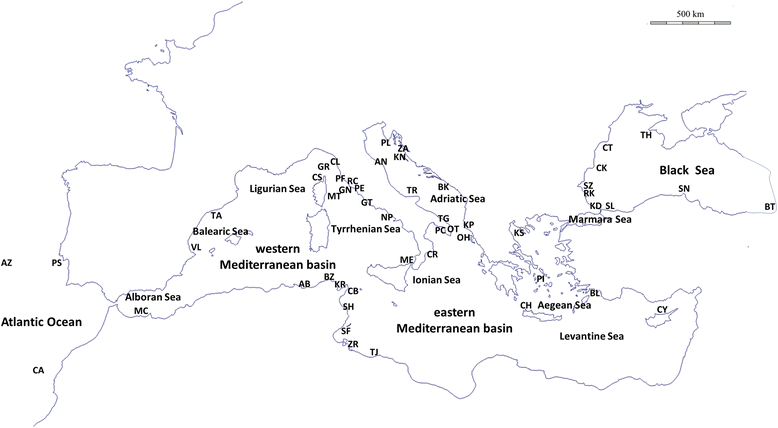
Background: The extent of genetic structure of a species is determined by the amount of current gene flow and the impact of historical and demographic factors. Most marine invertebrates have planktonic larvae and consequently wide potential dispersal, so that genetic uniformity should be common. However, phylogeographic investigations reveal that panmixia is rare in the marine realm. Phylogeographic patterns commonly coincide with geographic transitions acting as barriers to gene flow. In the Mediterranean Sea and adjoining areas, the best known barriers are the Atlantic-Mediterranean transition, the Siculo-Tunisian Strait and the boundary between Aegean and Black seas. Here, we perform the so far broadest phylogeographic analysis of the crab Pachygrapsus marmoratus, common across the north-eastern Atlantic Ocean, Mediterranean and Black seas. Previous studies revealed no or weak genetic structuring at meso-geographic scale based on mtDNA, while genetic heterogeneity at local scale was recorded with microsatellites, even if without clear geographic patterns. Continuing the search for phylogeographic signal, we here enlarge the mtDNA dataset including 51 populations and covering most of the species' distribution range.
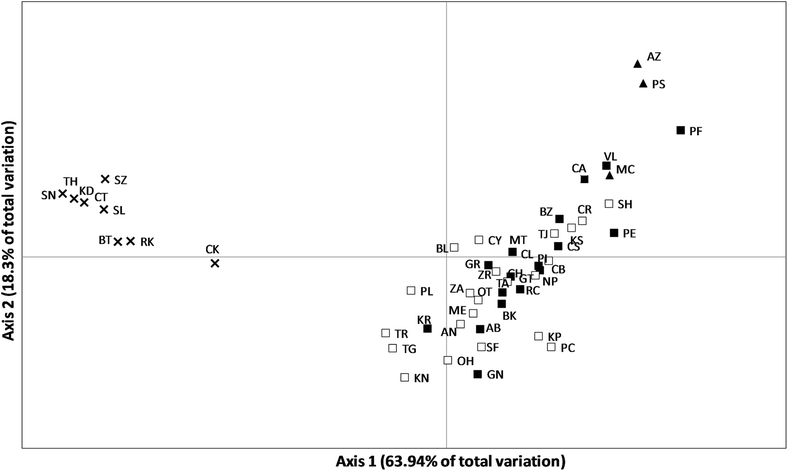
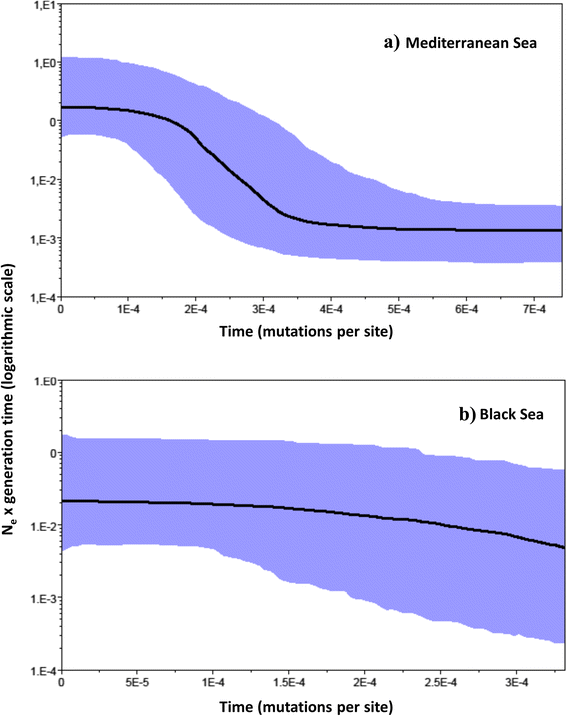
Results: This enlarged dataset provides new evidence of three genetically separable groups, corresponding to the Portuguese Atlantic Ocean, Mediterranean Sea plus Canary Islands, and Black Sea. Surprisingly, hierarchical AMOVA and Principal Coordinates Analysis agree that our Canary Islands population is closer to western Mediterranean populations than to mainland Portugal and Azores populations. Within the Mediterranean Sea, we record genetic homogeneity, suggesting that population connectivity is unaffected by the transition between the western and eastern Mediterranean. The Mediterranean metapopulation seems to have experienced a relatively recent expansion around 100,000 years ago.
Conclusions: Our results suggest that the phylogeographic pattern of P. marmoratus is shaped by the geological history of Mediterranean and adjacent seas, restricted current gene flow among different marginal seas, and incomplete lineage sorting. However, they also caution from exclusively testing well-known biogeographic barriers, thereby neglecting other possible phylogeographic patterns. Mostly, this study provides evidence that a geographically exhaustive dataset is necessary to detect shallow phylogeographic structure within widespread marine species with larval dispersal, questioning all studies where species have been categorized as panmictic based on numerically and geographically limited datasets.
Keywords: Crustacea Brachyura; Larval dispersal; Mediterranean Sea; Phylogeography; mtDNA CoxI.
Figures

Similar articles
-
Investigation of mechanisms underlying chaotic genetic patchiness in the intertidal marbled crab Pachygrapsus marmoratus (Brachyura: Grapsidae) across the Ligurian Sea.BMC Evol Biol. 2020 Aug 24;20(1):108. doi: 10.1186/s12862-020-01672-x. BMC Evol Biol. 2020. PMID: 32831022 Free PMC article.
-
Parapatric genetic divergence among deep evolutionary lineages in the Mediterranean green crab, Carcinus aestuarii (Brachyura, Portunoidea, Carcinidae), accounts for a sharp phylogeographic break in the Eastern Mediterranean.BMC Evol Biol. 2018 Apr 11;18(1):53. doi: 10.1186/s12862-018-1167-4. BMC Evol Biol. 2018. PMID: 29642852 Free PMC article.
-
Phylogeographic and evolutionary history analyses of the warty crab Eriphia verrucosa (Decapoda, Brachyura, Eriphiidae) unveil genetic imprints of a late Pleistocene vicariant event across the Gibraltar Strait, erased by postglacial expansion and admixture among refugial lineages.BMC Evol Biol. 2019 May 17;19(1):105. doi: 10.1186/s12862-019-1423-2. BMC Evol Biol. 2019. PMID: 31101005 Free PMC article.
-
Pillars of Hercules: is the Atlantic-Mediterranean transition a phylogeographical break?Mol Ecol. 2007 Nov;16(21):4426-44. doi: 10.1111/j.1365-294X.2007.03477.x. Epub 2007 Oct 1. Mol Ecol. 2007. PMID: 17908222 Review.
-
Harbour Porpoises, Phocoena phocoena, in the Mediterranean Sea and Adjacent Regions: Biogeographic Relicts of the Last Glacial Period.Adv Mar Biol. 2016;75:333-358. doi: 10.1016/bs.amb.2016.08.006. Epub 2016 Sep 28. Adv Mar Biol. 2016. PMID: 27770989 Review.
Cited by
-
Mitochondrial marker implies fishery separate management units for spotted sardinella, Amblygaster sirm (Walbaum, 1792) populations in the South China Sea and the Andaman Sea.PeerJ. 2022 Jul 15;10:e13706. doi: 10.7717/peerj.13706. eCollection 2022. PeerJ. 2022. PMID: 35860045 Free PMC article.
-
Mitonuclear genetic patterns of divergence in the marbled crab, Pachygrapsus marmoratus (Fabricius, 1787) along the Turkish seas.PLoS One. 2022 Apr 5;17(4):e0266506. doi: 10.1371/journal.pone.0266506. eCollection 2022. PLoS One. 2022. PMID: 35381029 Free PMC article.
-
Investigation of mechanisms underlying chaotic genetic patchiness in the intertidal marbled crab Pachygrapsus marmoratus (Brachyura: Grapsidae) across the Ligurian Sea.BMC Evol Biol. 2020 Aug 24;20(1):108. doi: 10.1186/s12862-020-01672-x. BMC Evol Biol. 2020. PMID: 32831022 Free PMC article.
-
Population genetic structure and evolutionary history of Bale monkeys (Chlorocebus djamdjamensis) in the southern Ethiopian Highlands.BMC Evol Biol. 2018 Jul 10;18(1):106. doi: 10.1186/s12862-018-1217-y. BMC Evol Biol. 2018. PMID: 29986642 Free PMC article.
-
Interannual fluctuations in connectivity among crab populations (Liocarcinus depurator) along the Atlantic-Mediterranean transition.Sci Rep. 2022 Jun 13;12(1):9797. doi: 10.1038/s41598-022-13941-4. Sci Rep. 2022. PMID: 35697727 Free PMC article.
References
-
- Bianchi NC, Morri C. Marine biodiversity of the Mediterranean Sea: situation, problems and prospects for future research. Mar Poll Bull. 2000;40:367–376. doi: 10.1016/S0025-326X(00)00027-8. - DOI
-
- Quignard JP. La Mediterranee, creuset ichthyologique. Bolletin Zoologica (Suppl II) 1978;840–45:23–36. doi: 10.1080/11250007809440264. - DOI
-
- Zane L, Ostellari L, Maccatrozzo L, Bargelloni L, Cuzin-Roudy J, Buchholz F, et al. Genetic differentiation in a pelagic crustacean (Meganyctiphanes norvegica Euphausiacea) from the North east Atlantic and the Mediterranean Sea. Mar Biol. 2000;136:191–199. doi: 10.1007/s002270050676. - DOI
-
- Papetti C, Zane L, Bortolotto E, Bucklin A, Patarnello T. Genetic differentiation and local temporal stability of population structure in the euphausiid Meganyctiphanes norvegica. Mar Ecol Progr Ser. 2005;289:225–235. doi: 10.3354/meps289225. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

