Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1
- PMID: 27457125
- PMCID: PMC5074603
- DOI: 10.1104/pp.16.00285
Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1
Abstract
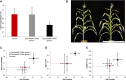
Flowering time (FTi) control is well examined in the long-day plant Arabidopsis (Arabidopsis thaliana), and increasing knowledge is available for the short-day plant rice (Oryza sativa). In contrast, little is known in the day-neutral and agronomically important crop plant maize (Zea mays). To learn more about FTi and to identify novel regulators in this species, we first compared the time points of floral transition of almost 30 maize inbred lines and show that tropical lines exhibit a delay in flowering transition of more than 3 weeks under long-day conditions compared with European flint lines adapted to temperate climate zones. We further analyzed the leaf transcriptomes of four lines that exhibit strong differences in flowering transition to identify new key players of the flowering control network in maize. We found strong differences among regulated genes between these lines and thus assume that the regulation of FTi is very complex in maize. Especially genes encoding MADS box transcriptional regulators are up-regulated in leaves during the meristem transition. ZmMADS1 was selected for functional studies. We demonstrate that it represents a functional ortholog of the central FTi integrator SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) of Arabidopsis. RNA interference-mediated down-regulation of ZmMADS1 resulted in a delay of FTi in maize, while strong overexpression caused an early-flowering phenotype, indicating its role as a flowering activator. Taken together, we report that ZmMADS1 represents a positive FTi regulator that shares an evolutionarily conserved function with SOC1 and may now serve as an ideal stating point to study the integration and variation of FTi pathways also in maize.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures

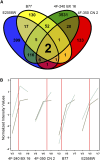

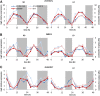
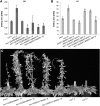
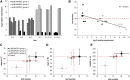
References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 - PubMed
-
- Abendroth LJ, Elmore RW, Boyer MJ, Marlay SK (2011) Corn Growth and Development. Iowa State University Extension, Ames, IA
-
- Andrés F, Porri A, Torti S, Mateos J, Romera-Branchat M, García-Martínez JL, Fornara F, Gregis V, Kater MM, Coupland G (2014) SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA 111: E2760–E2769 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

