The SaeRS Two-Component System Is a Direct and Dominant Transcriptional Activator of Toxic Shock Syndrome Toxin 1 in Staphylococcus aureus
- PMID: 27457715
- PMCID: PMC5019057
- DOI: 10.1128/JB.00425-16
The SaeRS Two-Component System Is a Direct and Dominant Transcriptional Activator of Toxic Shock Syndrome Toxin 1 in Staphylococcus aureus
Abstract
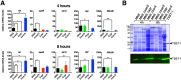
Toxic shock syndrome toxin 1 (TSST-1) is a Staphylococcus aureus superantigen that has been implicated in both menstrual and nonmenstrual toxic shock syndrome (TSS). Despite the important role of TSST-1 in severe human disease, a comprehensive understanding of staphylococcal regulatory factors that control TSST-1 expression remains incomplete. The S. aureus exotoxin expression (Sae) operon contains a well-characterized two-component system that regulates a number of important exotoxins in S. aureus, although regulation of TSST-1 by the Sae system has not been investigated. We generated a defined deletion mutant of the Sae histidine kinase sensor (saeS) in the prototypic menstrual TSS strain S. aureus MN8. Mutation of saeS resulted in a complete loss of TSST-1 expression. Using both luciferase reporter experiments and quantitative real-time PCR, we demonstrate that the Sae system is an important transcriptional activator of TSST-1 expression. Recombinant SaeR was able to bind directly to the tst promoter to a region containing two SaeR consensus binding sites. Although the stand-alone SarA transcriptional regulator has been shown to be both a positive and a negative regulator of TSST-1, deletion of sarA in S. aureus MN8 resulted in a dramatic overexpression of TSST-1. As expected, mutation of agr also reduced TSST-1 expression, but this phenotype appeared to be independent of Sae. A double mutation of saeS and sarA resulted in the loss of TSST-1 expression. This work indicates that the Sae system is a dominant and direct transcriptional activator that is required for expression of TSST-1.
Importance: The TSST-1 superantigen is an exotoxin, produced by some strains of S. aureus, that has a clear role in both menstrual and nonmenstrual TSS. Although the well-characterized agr quorum sensing system is a known positive regulator of TSST-1, the molecular mechanisms that directly control TSST-1 expression are only partially understood. Our studies demonstrate that the Sae two-component regulatory system is a positive transcriptional regulator that binds directly to the TSST-1 promoter, and furthermore, our data suggest that Sae is required for expression of TSST-1. This work highlights how major regulatory circuits can converge to fine-tune exotoxin expression and suggests that the Sae regulatory system may be an important target for antivirulence strategies.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


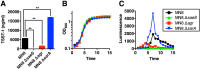



References
-
- Centers for Disease Control and Prevention. 2011. Toxic shock syndrome; 2011 case definition. Centers for Disease Control and Prevention, Atlanta, GA.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

