Computational investigation of cold denaturation in the Trp-cage miniprotein
- PMID: 27457961
- PMCID: PMC4987839
- DOI: 10.1073/pnas.1607500113
Computational investigation of cold denaturation in the Trp-cage miniprotein
Abstract
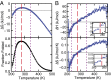
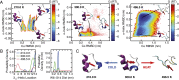
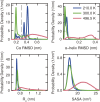
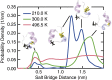
The functional native states of globular proteins become unstable at low temperatures, resulting in cold unfolding and impairment of normal biological function. Fundamental understanding of this phenomenon is essential to rationalizing the evolution of freeze-tolerant organisms and developing improved strategies for long-term preservation of biological materials. We present fully atomistic simulations of cold denaturation of an α-helical protein, the widely studied Trp-cage miniprotein. In contrast to the significant destabilization of the folded structure at high temperatures, Trp-cage cold denatures at 210 K into a compact, partially folded state; major elements of the secondary structure, including the α-helix, are conserved, but the salt bridge between aspartic acid and arginine is lost. The stability of Trp-cage's α-helix at low temperatures suggests a possible evolutionary explanation for the prevalence of such structures in antifreeze peptides produced by cold-weather species, such as Arctic char. Although the 310-helix is observed at cold conditions, its position is shifted toward Trp-cage's C-terminus. This shift is accompanied by intrusion of water into Trp-cage's interior and the hydration of buried hydrophobic residues. However, our calculations also show that the dominant contribution to the favorable energetics of low-temperature unfolding of Trp-cage comes from the hydration of hydrophilic residues.
Keywords: Trp-cage miniprotein; cold denaturation; protein folding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
A Computational Study of the Ionic Liquid-Induced Destabilization of the Miniprotein Trp-Cage.J Phys Chem B. 2018 May 31;122(21):5707-5715. doi: 10.1021/acs.jpcb.8b01722. Epub 2018 Apr 16. J Phys Chem B. 2018. PMID: 29617131
-
Computational Investigation of the Effect of Pressure on Protein Stability.J Phys Chem Lett. 2019 Apr 18;10(8):1894-1899. doi: 10.1021/acs.jpclett.9b00545. Epub 2019 Apr 4. J Phys Chem Lett. 2019. PMID: 30939023
-
Folding dynamics of the Trp-cage miniprotein: evidence for a native-like intermediate from combined time-resolved vibrational spectroscopy and molecular dynamics simulations.J Phys Chem B. 2013 Oct 3;117(39):11490-501. doi: 10.1021/jp404714c. Epub 2013 Sep 19. J Phys Chem B. 2013. PMID: 24050152
-
Folding dynamics of Trp-cage in the presence of chemical interference and macromolecular crowding. I.J Chem Phys. 2011 Nov 7;135(17):175101. doi: 10.1063/1.3656691. J Chem Phys. 2011. PMID: 22070323
-
The hydrophobic effect: a new insight from cold denaturation and a two-state water structure.Crit Rev Biochem Mol Biol. 2002;37(2):55-69. doi: 10.1080/10409230290771456. Crit Rev Biochem Mol Biol. 2002. PMID: 12027264 Review.
Cited by
-
Conformational Stability and Denaturation Processes of Proteins Investigated by Electrophoresis under Extreme Conditions.Molecules. 2022 Oct 13;27(20):6861. doi: 10.3390/molecules27206861. Molecules. 2022. PMID: 36296453 Free PMC article. Review.
-
Spatially resolved free-energy contributions of native fold and molten-globule-like Crambin.Biophys J. 2021 Aug 17;120(16):3470-3482. doi: 10.1016/j.bpj.2021.05.019. Epub 2021 Jun 2. Biophys J. 2021. PMID: 34087209 Free PMC article.
-
Ligand-induced cold activation of TRPV3.Res Sq [Preprint]. 2025 Jan 21:rs.3.rs-5759985. doi: 10.21203/rs.3.rs-5759985/v1. Res Sq. 2025. PMID: 39975930 Free PMC article. Preprint.
-
From Protein Design to the Energy Landscape of a Cold Unfolding Protein.J Phys Chem B. 2022 Feb 17;126(6):1212-1231. doi: 10.1021/acs.jpcb.1c10750. Epub 2022 Feb 7. J Phys Chem B. 2022. PMID: 35128921 Free PMC article.
-
Comparison of the Determination of Some Antihypertensive Drugs in Clinical Human Plasma Samples by Solvent Front Position Extraction and Precipitation Modes.Molecules. 2023 Feb 27;28(5):2213. doi: 10.3390/molecules28052213. Molecules. 2023. PMID: 36903457 Free PMC article.
References
-
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. - PubMed
-
- Zhang J, Peng X, Jonas A, Jonas J. NMR study of the cold, heat, and pressure unfolding of ribonuclease A. Biochemistry. 1995;34(27):8631–8641. - PubMed
-
- Privalov PL. Stability of proteins: Small globular proteins. Adv Protein Chem. 1979;33:167–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

