Structural Basis of Host Autophagy-related Protein 8 (ATG8) Binding by the Irish Potato Famine Pathogen Effector Protein PexRD54
- PMID: 27458016
- PMCID: PMC5025708
- DOI: 10.1074/jbc.M116.744995
Structural Basis of Host Autophagy-related Protein 8 (ATG8) Binding by the Irish Potato Famine Pathogen Effector Protein PexRD54
Abstract
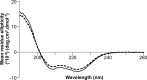
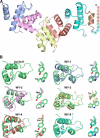
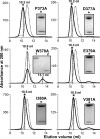
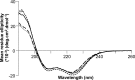
Filamentous plant pathogens deliver effector proteins to host cells to promote infection. The Phytophthora infestans RXLR-type effector PexRD54 binds potato ATG8 via its ATG8 family-interacting motif (AIM) and perturbs host-selective autophagy. However, the structural basis of this interaction remains unknown. Here, we define the crystal structure of PexRD54, which includes a modular architecture, including five tandem repeat domains, with the AIM sequence presented at the disordered C terminus. To determine the interface between PexRD54 and ATG8, we solved the crystal structure of potato ATG8CL in complex with a peptide comprising the effector's AIM sequence, and we established a model of the full-length PexRD54-ATG8CL complex using small angle x-ray scattering. Structure-informed deletion of the PexRD54 tandem domains reveals retention of ATG8CL binding in vitro and in planta This study offers new insights into structure/function relationships of oomycete RXLR effectors and how these proteins engage with host cell targets to promote disease.
Keywords: autophagy; effector protein; host-pathogen interaction; plant molecular biology; plant pathogen; protein structure; protein-protein interaction.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

