Pathogenesis and Phylogenetic Analyses of Two Avian Influenza H7N1 Viruses Isolated from Wild Birds
- PMID: 27458455
- PMCID: PMC4935687
- DOI: 10.3389/fmicb.2016.01066
Pathogenesis and Phylogenetic Analyses of Two Avian Influenza H7N1 Viruses Isolated from Wild Birds
Abstract
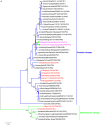
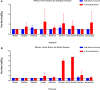
The emergence of human infections with a novel H7N9 influenza strain has raised global concerns about a potential human pandemic. To further understand the character of other influenza viruses of the H7 subtype, we selected two H7N1 avian influenza viruses (AIVs) isolated from wild birds during routine surveillance in China: A/Baer's Pochard/Hunan/414/2010 (BP/HuN/414/10) (H7N1) and A/Common Pochard/Xianghai/420/2010 (CP/XH/420/10) (H7N1). To better understand the molecular characteristics of these two isolated H7N1 viruses, we sequenced and phylogenetically analyzed their entire genomes. The results showed that the two H7N1 strains belonged to a Eurasian branch, originating from a common ancestor. Phylogenetic analysis of their hemagglutinin (HA) genes showed that BP/HuN/414/10 and CP/XH/420/10 have a more distant genetic relationship with A/Shanghai/13/2013 (H7N9), with similarities of 91.6 and 91.4%, respectively. To assess the replication and pathogenicity of these viruses in different hosts, they were inoculated in chickens, ducks and mice. Although, both CP/XH/420/10 and BP/HuN/414/10 can infect chickens, ducks and mice, they exhibited different replication capacities in these animals. The results of this study demonstrated that two low pathogenic avian influenza (LPAI) H7N1 viruses of the Eurasian branch could infect mammals and may even have the potential to infect humans. Therefore, it is important to monitor H7 viruses in both domestic and wild birds.
Keywords: Avian influenza virus; H7N1; H7N9; pathogenic analyses; phylogenetic analysis.
Figures








Similar articles
-
Replication and pathogenic potential of influenza A virus subtypes H3, H7, and H15 from free-range ducks in Bangladesh in mammals.Emerg Microbes Infect. 2018 Apr 25;7(1):70. doi: 10.1038/s41426-018-0072-7. Emerg Microbes Infect. 2018. PMID: 29691394 Free PMC article.
-
Low pathogenic H7 subtype avian influenza viruses isolated from domestic ducks in South Korea and the close association with isolates of wild birds.J Gen Virol. 2012 Jun;93(Pt 6):1278-1287. doi: 10.1099/vir.0.041269-0. Epub 2012 Mar 14. J Gen Virol. 2012. PMID: 22422062
-
Deletion of the C-terminal ESEV domain of NS1 does not affect the replication of a low-pathogenic avian influenza virus H7N1 in ducks and chickens.J Gen Virol. 2013 Jan;94(Pt 1):50-58. doi: 10.1099/vir.0.045153-0. Epub 2012 Oct 10. J Gen Virol. 2013. PMID: 23052391
-
Genetic relationship of H3 subtype avian influenza viruses isolated from domestic ducks and wild birds in Korea and their pathogenic potential in chickens and ducks.Vet Microbiol. 2012 Mar 23;155(2-4):147-57. doi: 10.1016/j.vetmic.2011.08.028. Epub 2011 Sep 3. Vet Microbiol. 2012. PMID: 21955449
-
Emergence and Adaptation of a Novel Highly Pathogenic H7N9 Influenza Virus in Birds and Humans from a 2013 Human-Infecting Low-Pathogenic Ancestor.J Virol. 2018 Jan 2;92(2):e00921-17. doi: 10.1128/JVI.00921-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29070694 Free PMC article.
Cited by
-
Mammalian pathogenicity and transmissibility of low pathogenic avian influenza H7N1 and H7N3 viruses isolated from North America in 2018.Emerg Microbes Infect. 2020 Dec;9(1):1037-1045. doi: 10.1080/22221751.2020.1764396. Emerg Microbes Infect. 2020. PMID: 32449503 Free PMC article.
-
Pathogenicity and Transmissibility of Clade 2.3.4.4h H5N6 Avian Influenza Viruses in Mammals.Animals (Basel). 2022 Nov 9;12(22):3079. doi: 10.3390/ani12223079. Animals (Basel). 2022. PMID: 36428307 Free PMC article.
-
Isolation and characterization of novel reassortant H6N1 avian influenza viruses from chickens in Eastern China.Virol J. 2018 Oct 24;15(1):164. doi: 10.1186/s12985-018-1063-y. Virol J. 2018. PMID: 30355336 Free PMC article.
-
Study of the host specificity of PB1-F2-associated virulence.Virulence. 2021 Dec;12(1):1647-1660. doi: 10.1080/21505594.2021.1933848. Virulence. 2021. PMID: 34125653 Free PMC article.
-
Host Range of Influenza A Virus H1 to H16 in Eurasian Ducks Based on Tissue and Receptor Binding Studies.J Virol. 2021 Feb 24;95(6):e01873-20. doi: 10.1128/JVI.01873-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361418 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

