Reprogramming, Circular Reasoning and Self versus Non-self: One-Stop Shopping with RNA Editing
- PMID: 27458478
- PMCID: PMC4937755
- DOI: 10.3389/fgene.2016.00100
Reprogramming, Circular Reasoning and Self versus Non-self: One-Stop Shopping with RNA Editing
Abstract
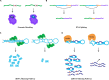
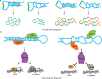
Transcription of genetic information from archival DNA into RNA molecule working copies is vital for proper cellular function and is highly accurate. In turn, RNAs serve structural, enzymatic, and regulatory roles, as well as being informational templates for the ribosomal translation of proteins. Following RNA synthesis, maturing of RNA molecules occurs through various RNA processing events. One component of the collection of processes involving RNA species, broadly defined as RNA metabolism, is the RNA-editing pathway and is found in all animals. Acting specifically on RNA substrates with double-stranded character, RNA editing has been shown to regulate a plethora of genomic outputs, including gene recoding, RNA splicing, biogenesis and targeting actions of microRNAs and small interfering RNAs, and global gene expression. Recent evidence suggests that RNA modifications mediated via RNA editing influence the biogenesis of circular RNAs and safeguard against aberrant innate immune responses generated to endogenous RNA sources. These novel roles have the potential to contribute new insights into molecular mechanisms underlying pathogenesis mediated by mishandling of double-stranded RNA. Here, we discuss recent advances in the field, which highlight novel roles associated with the RNA-editing process and emphasize their importance during cellular RNA metabolism. In addition, we highlight the relevance of these newly discovered roles in the context of neurological disorders and the more general concept of innate recognition of self versus non-self.
Keywords: RNA editing; RNA metabolism; RNA silencing; RNA splicing; autoimmune disease; circular RNAs; immune responses; neurological disorders.
Figures
Similar articles
-
A-to-I RNA Editing: Current Knowledge Sources and Computational Approaches with Special Emphasis on Non-Coding RNA Molecules.Front Bioeng Biotechnol. 2015 Mar 25;3:37. doi: 10.3389/fbioe.2015.00037. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 25859542 Free PMC article. Review.
-
ADAR-mediated RNA editing in non-coding RNA sequences.Sci China Life Sci. 2013 Oct;56(10):944-52. doi: 10.1007/s11427-013-4546-5. Epub 2013 Sep 5. Sci China Life Sci. 2013. PMID: 24008387 Review.
-
All I's on the RADAR: role of ADAR in gene regulation.FEBS Lett. 2018 Sep;592(17):2860-2873. doi: 10.1002/1873-3468.13093. Epub 2018 May 25. FEBS Lett. 2018. PMID: 29770436 Free PMC article. Review.
-
Posttranscriptional recoding by RNA editing.Adv Protein Chem Struct Biol. 2012;86:193-224. doi: 10.1016/B978-0-12-386497-0.00006-2. Adv Protein Chem Struct Biol. 2012. PMID: 22243585 Review.
-
Functions of the RNA Editing Enzyme ADAR1 and Their Relevance to Human Diseases.Genes (Basel). 2016 Dec 17;7(12):129. doi: 10.3390/genes7120129. Genes (Basel). 2016. PMID: 27999332 Free PMC article. Review.
Cited by
-
The C. elegans neural editome reveals an ADAR target mRNA required for proper chemotaxis.Elife. 2017 Sep 19;6:e28625. doi: 10.7554/eLife.28625. Elife. 2017. PMID: 28925356 Free PMC article.
-
Testicular adenosine to inosine RNA editing in the mouse is mediated by ADARB1.Biol Reprod. 2017 Jan 1;96(1):244-253. doi: 10.1095/biolreprod.116.145151. Biol Reprod. 2017. PMID: 28395340 Free PMC article.
-
Predicting RNA hyper-editing with a novel tool when unambiguous alignment is impossible.BMC Genomics. 2017 Jul 10;18(1):522. doi: 10.1186/s12864-017-3898-9. BMC Genomics. 2017. PMID: 28693467 Free PMC article.
-
RNA editing in cancer impacts mRNA abundance in immune response pathways.Genome Biol. 2020 Oct 26;21(1):268. doi: 10.1186/s13059-020-02171-4. Genome Biol. 2020. PMID: 33106178 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

