Feature Selection Methods for Early Predictive Biomarker Discovery Using Untargeted Metabolomic Data
- PMID: 27458587
- PMCID: PMC4937038
- DOI: 10.3389/fmolb.2016.00030
Feature Selection Methods for Early Predictive Biomarker Discovery Using Untargeted Metabolomic Data
Abstract
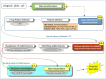
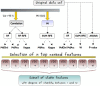
Untargeted metabolomics is a powerful phenotyping tool for better understanding biological mechanisms involved in human pathology development and identifying early predictive biomarkers. This approach, based on multiple analytical platforms, such as mass spectrometry (MS), chemometrics and bioinformatics, generates massive and complex data that need appropriate analyses to extract the biologically meaningful information. Despite various tools available, it is still a challenge to handle such large and noisy datasets with limited number of individuals without risking overfitting. Moreover, when the objective is focused on the identification of early predictive markers of clinical outcome, few years before occurrence, it becomes essential to use the appropriate algorithms and workflow to be able to discover subtle effects among this large amount of data. In this context, this work consists in studying a workflow describing the general feature selection process, using knowledge discovery and data mining methodologies to propose advanced solutions for predictive biomarker discovery. The strategy was focused on evaluating a combination of numeric-symbolic approaches for feature selection with the objective of obtaining the best combination of metabolites producing an effective and accurate predictive model. Relying first on numerical approaches, and especially on machine learning methods (SVM-RFE, RF, RF-RFE) and on univariate statistical analyses (ANOVA), a comparative study was performed on an original metabolomic dataset and reduced subsets. As resampling method, LOOCV was applied to minimize the risk of overfitting. The best k-features obtained with different scores of importance from the combination of these different approaches were compared and allowed determining the variable stabilities using Formal Concept Analysis. The results revealed the interest of RF-Gini combined with ANOVA for feature selection as these two complementary methods allowed selecting the 48 best candidates for prediction. Using linear logistic regression on this reduced dataset enabled us to obtain the best performances in terms of prediction accuracy and number of false positive with a model including 5 top variables. Therefore, these results highlighted the interest of feature selection methods and the importance of working on reduced datasets for the identification of predictive biomarkers issued from untargeted metabolomics data.
Keywords: biomarker discovery; feature selection; formal concept analysis; machine learning; metabolomics; prediction; univariate statistics; visualization.
Figures

References
-
- Agrawal R., Mielinski T., Swami A. (eds.). (1993). Mining association rules between sets of items in large databasesMining association rules between sets of items in large databases, in ACM SIGMOD Conference (Washington, DC: ).
-
- Biau G. (2012). Analysis of a random forests model. J. Mach. Learn. Res. 13, 1063–1095.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

