Coordinate regulation of autophagy and the ubiquitin proteasome system by MTOR
- PMID: 27459110
- PMCID: PMC5079674
- DOI: 10.1080/15548627.2016.1205770
Coordinate regulation of autophagy and the ubiquitin proteasome system by MTOR
Abstract
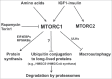
Proteins in eukaryotic cells are continually being degraded to amino acids either by the ubiquitin proteasome system (UPS) or by the autophagic-lysosomal pathway. The breakdown of proteins by these 2 degradative pathways involves totally different enzymes that function in distinct subcellular compartments. While most studies of the UPS have focused on the selective ubiquitination and breakdown of specific cell proteins, macroautophagy/autophagy is a more global nonselective process. Consequently, the UPS and autophagy were traditionally assumed to serve distinct physiological functions and to be regulated in quite different manners. However, recent findings indicate that protein breakdown by these 2 systems is coordinately regulated by important physiological stimuli. The activation of MTORC1 by nutrients and hormones rapidly suppresses proteolysis by both proteasomes and autophagy, which helps promote protein accumulation, whereas in nutrient-poor conditions, MTORC1 inactivation causes the simultaneous activation of these 2 degradative pathways to supply the deprived cells with a source of amino acids. Also this selective breakdown of key anabolic proteins by the UPS upon MTORC1 inhibition can help limit growth-related processes (e.g., cholesterol biosynthesis). Thus, the collaboration of these 2 degradative systems, together with the simultaneous control of protein translation by MTORC1, provide clear advantages to the organism in both growth and starvation conditions.
Keywords: MTOR; autophagy; proteasomes; ubiquitination.
Figures
References
-
- Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-Sensing Mechanisms across Evolution. Cell 2015; 161:67-83; PMID:25815986; http://dx.doi.org/10.1016/j.cell.2015.02.041 - DOI - PMC - PubMed
-
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109-13; PMID:22552098; http://dx.doi.org/10.1038/nature11083 - DOI - PMC - PubMed
-
- Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol 2015; 25:354-63; PMID:25759175; http://dx.doi.org/10.1016/j.tcb.2015.02.002 - DOI - PMC - PubMed
-
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994; 78:761-71; PMID:8087844; http://dx.doi.org/10.1016/S0092-8674(94)90462-6 - DOI - PubMed
-
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007; 6:472-83; PMID:18054316; http://dx.doi.org/10.1016/j.cmet.2007.11.004 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
