Role of Cell Membrane-Vector Interactions in Successful Gene Delivery
- PMID: 27459207
- PMCID: PMC12459993
- DOI: 10.1021/acs.accounts.6b00200
Role of Cell Membrane-Vector Interactions in Successful Gene Delivery
Abstract
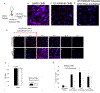
Cationic polymers have been investigated as nonviral vectors for gene delivery due to their favorable safety profile when compared to viral vectors. However, nonviral vectors are limited by poor efficacy in inducing gene expression. The physicochemical properties of cationic polymers enabling successful gene expression have been investigated in order to improve expression efficiency and safety. Studies over the past several years have focused on five possible rate-limiting processes to explain the differences in gene expression: (1) endosomal release, (2) transport within specific intracellular pathways, (3) protection of DNA from nucleases, (4) transport into the nucleus, and (5) DNA release from vectors. However, determining the relative importance of these processes and the vector properties necessary for optimization remain a challenge to the field. In this Account, we describe over a decade of studies focused on understanding the interaction of cationic polymer and cationic polymer/oligonucleotide (polyplex) interactions with model lipid membranes, cell membranes, and cells in culture. In particular, we have been interested in how the interaction between cationic polymers and the membrane influences the intracellular transport of intact DNA to the nucleus. Recent advances in microfluidic patch clamp techniques enabled us to quantify polyplex cell membrane interactions at the cellular level with precise control over material concentrations and exposure times. In attempting to relate these findings to subsequent intracellular transport of DNA and expression of protein, we needed to develop an approach that could distinguish DNA that was intact and potentially functional for gene expression from the much larger pool of degraded, nonfunctional DNA within the cell. We addressed this need by developing a FRET oligonucleotide molecular beacon (OMB) to monitor intact DNA transport. The research highlighted in this Account builds to the conclusion that polyplex transported DNA is released from endosomes by free cationic polymer intercalated into the endosomal membrane. This cationic polymer initially interacts with the cell plasma membrane and appears to reach the endosome by lipid cycling mechanisms. The fraction of cells displaying release of intact DNA from endosomes quantitatively predicts the fraction of cells displaying gene expression for both linear poly(ethylenimine) (L-PEI; an effective vector) and generation five poly(amidoamine) dendrimer (G5 PAMAM; an ineffective vector). Moreover, intact OMB delivered with G5 PAMAM, which normally is confined to endosomes, was released by the subsequent addition of L-PEI with a corresponding 10-fold increase in transgene expression. These observations are consistent with experiments demonstrating that cationic polymer/membrane partition coefficients, not polyplex/membrane partition coefficients, predict successful gene expression. Interestingly, a similar partitioning of cationic polymers into the mitochondrial membranes has been proposed to explain the cytotoxicity of these materials. Thus, the proposed model indicates the same physicochemical property (partitioning into lipid bilayers) is linked to release from endosomes, giving protein expression, and to cytotoxicity.
Figures

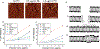


References
-
- Griesenbach U; Alton EWFW Gene Transfer to the Lung: Lessons Learned from More than 2 Decades of CF Gene Therapy. Adv. Drug Deliv. Rev. 2009, 61, 128–139. - PubMed
-
- Gori JL; Hsu PD; Maeder ML; Shen S; Welstead GG; Bumcrot D Delivery and Specificity of CRISPR/Cas9 Genome Editing Technologies for Human Gene Therapy. Hum. Gene Ther. 2015, 26, 443–451. - PubMed
-
- Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 2014, 15, 541–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

