An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients
- PMID: 27460420
- PMCID: PMC5117943
- DOI: 10.1038/ejhg.2016.99
An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients
Abstract
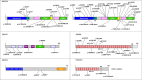
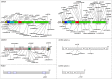
Usher syndrome (USH), the most prevalent cause of hereditary deafness-blindness, is an autosomal recessive and genetically heterogeneous disorder. Three clinical subtypes (USH1-3) are distinguishable based on the severity of the sensorineural hearing impairment, the presence or absence of vestibular dysfunction, and the age of onset of the retinitis pigmentosa. A total of 10 causal genes, 6 for USH1, 3 for USH2, and 1 for USH3, and an USH2 modifier gene, have been identified. A robust molecular diagnosis is required not only to improve genetic counseling, but also to advance gene therapy in USH patients. Here, we present an improved diagnostic strategy that is both cost- and time-effective. It relies on the sequential use of three different techniques to analyze selected genomic regions: targeted exome sequencing, comparative genome hybridization, and quantitative exon amplification. We screened a large cohort of 427 patients (139 USH1, 282 USH2, and six of undefined clinical subtype) from various European medical centers for mutations in all USH genes and the modifier gene. We identified a total of 421 different sequence variants predicted to be pathogenic, about half of which had not been previously reported. Remarkably, we detected large genomic rearrangements, most of which were novel and unique, in 9% of the patients. Thus, our strategy led to the identification of biallelic and monoallelic mutations in 92.7% and 5.8% of the USH patients, respectively. With an overall 98.5% mutation characterization rate, the diagnosis efficiency was substantially improved compared with previously reported methods.
Figures


References
-
- Boughman JA, Vernon M, Shaver KA: Usher syndrome: definition and estimate of prevalence from two high-risk populations. J Chronic Dis 1983; 36: 595–603. - PubMed
-
- Bonnet C, El-Amraoui A: Usher syndrome (sensorineural deafness and retinitis pigmentosa): pathogenesis, molecular diagnosis and therapeutic approaches. Curr Opin Neurol 2012; 25: 42–49. - PubMed
-
- Khateb S, Zelinger L, Mizrahi-Meissonnier L et al: A homozygous nonsense CEP250 mutation combined with a heterozygous nonsense C2orf71 mutation is associated with atypical Usher syndrome. J Med Genet 2014; 51: 460–469. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

