Novel HDAd/EBV Reprogramming Vector and Highly Efficient Ad/CRISPR-Cas Sickle Cell Disease Gene Correction
- PMID: 27460639
- PMCID: PMC4961958
- DOI: 10.1038/srep30422
Novel HDAd/EBV Reprogramming Vector and Highly Efficient Ad/CRISPR-Cas Sickle Cell Disease Gene Correction
Abstract
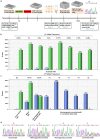
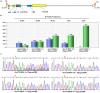
CRISPR/Cas enhanced correction of the sickle cell disease (SCD) genetic defect in patient-specific induced Pluripotent Stem Cells (iPSCs) provides a potential gene therapy for this debilitating disease. An advantage of this approach is that corrected iPSCs that are free of off-target modifications can be identified before differentiating the cells into hematopoietic progenitors for transplantation. In order for this approach to be practical, iPSC generation must be rapid and efficient. Therefore, we developed a novel helper-dependent adenovirus/Epstein-Barr virus (HDAd/EBV) hybrid reprogramming vector, rCLAE-R6, that delivers six reprogramming factors episomally. HDAd/EBV transduction of keratinocytes from SCD patients resulted in footprint-free iPSCs with high efficiency. Subsequently, the sickle mutation was corrected by delivering CRISPR/Cas9 with adenovirus followed by nucleoporation with a 70 nt single-stranded oligodeoxynucleotide (ssODN) correction template. Correction efficiencies of up to 67.9% (β(A)/[β(S)+β(A)]) were obtained. Whole-genome sequencing (WGS) of corrected iPSC lines demonstrated no CRISPR/Cas modifications in 1467 potential off-target sites and no modifications in tumor suppressor genes or other genes associated with pathologies. These results demonstrate that adenoviral delivery of reprogramming factors and CRISPR/Cas provides a rapid and efficient method of deriving gene-corrected, patient-specific iPSCs for therapeutic applications.
Figures

References
-
- Ingram V. M. A Specific Chemical Difference Between the Globins of Normal Human and Sickle-Cell Anæmia Hæmoglobin. Nature 178, 792- 794 (1956). - PubMed
-
- Ingram V. M. Gene Mutations in Human Hæmoglobin: the Chemical Difference Between Normal and Sickle Cell Hæmoglobin. Nature 180, 326- 328 (1957). - PubMed
-
- Thomas E. D. & Blume K. G. Historical markers in the development of allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant 5, 341–6 (1999). - PubMed
-
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–76 (2006). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

