A novel roseobacter phage possesses features of podoviruses, siphoviruses, prophages and gene transfer agents
- PMID: 27460944
- PMCID: PMC4961962
- DOI: 10.1038/srep30372
A novel roseobacter phage possesses features of podoviruses, siphoviruses, prophages and gene transfer agents
Abstract
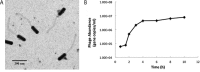
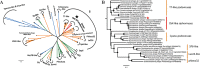
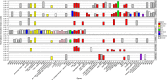
Bacteria in the Roseobacter lineage have been studied extensively due to their significant biogeochemical roles in the marine ecosystem. However, our knowledge on bacteriophage which infects the Roseobacter clade is still very limited. Here, we report a new bacteriophage, phage DSS3Φ8, which infects marine roseobacter Ruegeria pomeroyi DSS-3. DSS3Φ8 is a lytic siphovirus. Genomic analysis showed that DSS3Φ8 is most closely related to a group of siphoviruses, CbK-like phages, which infect freshwater bacterium Caulobacter crescentus. DSS3Φ8 contains a smaller capsid and has a reduced genome size (146 kb) compared to the CbK-like phages (205-279 kb). DSS3Φ8 contains the DNA polymerase gene which is closely related to T7-like podoviruses. DSS3Φ8 also contains the integrase and repressor genes, indicating its potential to involve in lysogenic cycle. In addition, four GTA (gene transfer agent) genes were identified in the DSS3Φ8 genome. Genomic analysis suggests that DSS3Φ8 is a highly mosaic phage that inherits the genetic features from siphoviruses, podoviruses, prophages and GTAs. This is the first report of CbK-like phages infecting marine bacteria. We believe phage isolation is still a powerful tool that can lead to discovery of new phages and help interpret the overwhelming unknown sequences in the viral metagenomics.
Figures



Similar articles
-
A newly isolated roseophage represents a distinct member of Siphoviridae family.Virol J. 2019 Nov 6;16(1):128. doi: 10.1186/s12985-019-1241-6. Virol J. 2019. PMID: 31694663 Free PMC article.
-
Complete genome sequence of the siphovirus Roseophage RDJLΦ 2 infecting Roseobacter denitrificans OCh114.Mar Genomics. 2016 Feb;25:17-19. doi: 10.1016/j.margen.2015.10.009. Epub 2015 Nov 2. Mar Genomics. 2016. PMID: 26541473
-
Characterization and Genomic Analysis of ValSw3-3, a New Siphoviridae Bacteriophage Infecting Vibrio alginolyticus.J Virol. 2020 May 4;94(10):e00066-20. doi: 10.1128/JVI.00066-20. Print 2020 May 4. J Virol. 2020. PMID: 32132234 Free PMC article.
-
Bacteriophages that infect marine roseobacters: genomics and ecology.Environ Microbiol. 2019 Jun;21(6):1885-1895. doi: 10.1111/1462-2920.14504. Epub 2019 Jan 13. Environ Microbiol. 2019. PMID: 30556267 Review.
-
Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas?ISME J. 2008 Jun;2(6):579-89. doi: 10.1038/ismej.2008.35. ISME J. 2008. PMID: 18521076 Review.
Cited by
-
Machine-Learning Classification Suggests That Many Alphaproteobacterial Prophages May Instead Be Gene Transfer Agents.Genome Biol Evol. 2019 Oct 1;11(10):2941-2953. doi: 10.1093/gbe/evz206. Genome Biol Evol. 2019. PMID: 31560374 Free PMC article.
-
Analyses of four new Caulobacter Phicbkviruses indicate independent lineages.J Gen Virol. 2019 Feb;100(2):321-331. doi: 10.1099/jgv.0.001218. Epub 2019 Jan 18. J Gen Virol. 2019. PMID: 30657445 Free PMC article.
-
Interactions and evolutionary relationships among bacterial mobile genetic elements.Nat Rev Microbiol. 2025 Jul;23(7):423-438. doi: 10.1038/s41579-025-01157-y. Epub 2025 Mar 11. Nat Rev Microbiol. 2025. PMID: 40069292 Review.
-
Amino Acid and Sugar Catabolism in the Marine Bacterium Phaeobacter inhibens DSM 17395 from an Energetic Viewpoint.Appl Environ Microbiol. 2019 Nov 27;85(24):e02095-19. doi: 10.1128/AEM.02095-19. Print 2019 Dec 15. Appl Environ Microbiol. 2019. PMID: 31604772 Free PMC article.
-
Vibrio Phage KVP40 Encodes a Functional NAD+ Salvage Pathway.J Bacteriol. 2017 Apr 11;199(9):e00855-16. doi: 10.1128/JB.00855-16. Print 2017 May 1. J Bacteriol. 2017. PMID: 28167526 Free PMC article.
References
-
- Brinkhoff T., Giebel H. A. & Simon M. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189, 531–539 (2008). - PubMed
-
- Moran M. A., Reisch C. R., Kiene R. P. & Whitman W. B. Genomic insights into bacterial DMSP transformations. Ann. Rev. Mar. Sci. 4, 523–542 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

