Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis
- PMID: 27461509
- PMCID: PMC4961963
- DOI: 10.1038/srep30379
Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis
Abstract
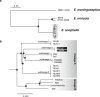
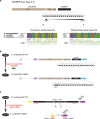
Elizabethkingia anophelis is an emerging pathogen involved in human infections and outbreaks in distinct world regions. We investigated the phylogenetic relationships and pathogenesis-associated genomic features of two neonatal meningitis isolates isolated 5 years apart from one hospital in Central African Republic and compared them with Elizabethkingia from other regions and sources. Average nucleotide identity firmly confirmed that E. anophelis, E. meningoseptica and E. miricola represent demarcated genomic species. A core genome multilocus sequence typing scheme, broadly applicable to Elizabethkingia species, was developed and made publicly available (http://bigsdb.pasteur.fr/elizabethkingia). Phylogenetic analysis revealed distinct E. anophelis sublineages and demonstrated high genetic relatedness between the African isolates, compatible with persistence of the strain in the hospital environment. CRISPR spacer variation between the African isolates was mirrored by the presence of a large mobile genetic element. The pan-genome of E. anophelis comprised 6,880 gene families, underlining genomic heterogeneity of this species. African isolates carried unique resistance genes acquired by horizontal transfer. We demonstrated the presence of extensive variation of the capsular polysaccharide synthesis gene cluster in E. anophelis. Our results demonstrate the dynamic evolution of this emerging pathogen and the power of genomic approaches for Elizabethkingia identification, population biology and epidemiology.
Figures



Similar articles
-
In Silico Identification of Three Types of Integrative and Conjugative Elements in Elizabethkingia anophelis Strains Isolated from around the World.mSphere. 2019 Apr 3;4(2):e00040-19. doi: 10.1128/mSphere.00040-19. mSphere. 2019. PMID: 30944210 Free PMC article.
-
The Evolutionary Trend and Genomic Features of an Emerging Lineage of Elizabethkingia anophelis Strains in Taiwan.Microbiol Spectr. 2022 Feb 23;10(1):e0168221. doi: 10.1128/spectrum.01682-21. Epub 2022 Jan 19. Microbiol Spectr. 2022. PMID: 35044198 Free PMC article.
-
Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis.Genome Biol Evol. 2014 May 6;6(5):1158-65. doi: 10.1093/gbe/evu094. Genome Biol Evol. 2014. PMID: 24803570 Free PMC article.
-
Mini review: New pathogen profiles: Elizabethkingia anophelis.Diagn Microbiol Infect Dis. 2017 Jun;88(2):201-205. doi: 10.1016/j.diagmicrobio.2017.03.007. Epub 2017 Mar 16. Diagn Microbiol Infect Dis. 2017. PMID: 28342565 Review.
-
[Elizabethkingia spp. infections in a university hospital: coinfection with SARS-CoV-2 and first report of Elizabethkingia anophelis in Chile].Rev Chilena Infectol. 2021 Oct;38(5):613-621. doi: 10.4067/s0716-10182021000500613. Rev Chilena Infectol. 2021. PMID: 35506827 Review. Spanish.
Cited by
-
Core-Proteomics-Based Annotation of Antigenic Targets and Reverse-Vaccinology-Assisted Design of Ensemble Immunogen against the Emerging Nosocomial Infection-Causing Bacterium Elizabethkingia meningoseptica.Int J Environ Res Public Health. 2021 Dec 24;19(1):194. doi: 10.3390/ijerph19010194. Int J Environ Res Public Health. 2021. PMID: 35010455 Free PMC article.
-
Structural characterization of a GNAT family acetyltransferase from Elizabethkingia anophelis bound to acetyl-CoA reveals a new dimeric interface.Sci Rep. 2021 Jan 14;11(1):1274. doi: 10.1038/s41598-020-79649-5. Sci Rep. 2021. PMID: 33446675 Free PMC article.
-
Insights from the draft genome into the pathogenicity of a clinical isolate of Elizabethkingia meningoseptica Em3.Stand Genomic Sci. 2017 Sep 16;12:56. doi: 10.1186/s40793-017-0269-8. eCollection 2017. Stand Genomic Sci. 2017. PMID: 28932346 Free PMC article.
-
MBLs, Rather Than Efflux Pumps, Led to Carbapenem Resistance in Fosfomycin and Aztreonam/Avibactam Resistant Elizabethkingia anophelis.Infect Drug Resist. 2021 Jan 29;14:315-327. doi: 10.2147/IDR.S294149. eCollection 2021. Infect Drug Resist. 2021. PMID: 33551643 Free PMC article.
-
Unraveling the global landscape of Elizabethkingia antibiotic resistance: A systematic review and meta-analysis.PLoS One. 2025 May 30;20(5):e0323313. doi: 10.1371/journal.pone.0323313. eCollection 2025. PLoS One. 2025. PMID: 40446068 Free PMC article.
References
-
- Lindh J. M., Borg-Karlson A. K. & Faye I. Transstadial and horizontal transfer of bacteria within a colony of Anopheles gambiae and oviposition response to bacteria-containing water. Acta Trop 107, 242–250 (2008). - PubMed
-
- Bloch K. C., Nadarajah R. & Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine 76, 30–41 (1997). - PubMed
-
- Güngör S., Ozen M., Akinci A. & Durmaz R. A. Chryseobacterium meningosepticum outbreak in a neonatal ward. Infect Control Hosp Epidemiol 24, 613–617 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

