A Novel Bifunctional Alkylphenol Anesthetic Allows Characterization of γ-Aminobutyric Acid, Type A (GABAA), Receptor Subunit Binding Selectivity in Synaptosomes
- PMID: 27462076
- PMCID: PMC5034043
- DOI: 10.1074/jbc.M116.736975
A Novel Bifunctional Alkylphenol Anesthetic Allows Characterization of γ-Aminobutyric Acid, Type A (GABAA), Receptor Subunit Binding Selectivity in Synaptosomes
Abstract
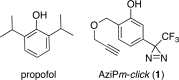
Propofol, an intravenous anesthetic, is a positive modulator of the GABAA receptor, but the mechanistic details, including the relevant binding sites and alternative targets, remain disputed. Here we undertook an in-depth study of alkylphenol-based anesthetic binding to synaptic membranes. We designed, synthesized, and characterized a chemically active alkylphenol anesthetic (2-((prop-2-yn-1-yloxy)methyl)-5-(3-(trifluoromethyl)-3H-diazirin-3-yl)phenol, AziPm-click (1)), for affinity-based protein profiling (ABPP) of propofol-binding proteins in their native state within mouse synaptosomes. The ABPP strategy captured ∼4% of the synaptosomal proteome, including the unbiased capture of five α or β GABAA receptor subunits. Lack of γ2 subunit capture was not due to low abundance. Consistent with this, independent molecular dynamics simulations with alchemical free energy perturbation calculations predicted selective propofol binding to interfacial sites, with higher affinities for α/β than γ-containing interfaces. The simulations indicated hydrogen bonding is a key component leading to propofol-selective binding within GABAA receptor subunit interfaces, with stable hydrogen bonds observed between propofol and α/β cavity residues but not γ cavity residues. We confirmed this by introducing a hydrogen bond-null propofol analogue as a protecting ligand for targeted-ABPP and observed a lack of GABAA receptor subunit protection. This investigation demonstrates striking interfacial GABAA receptor subunit selectivity in the native milieu, suggesting that asymmetric occupancy of heteropentameric ion channels by alkylphenol-based anesthetics is sufficient to induce modulation of activity.
Keywords: GABA receptor; anesthesia; anesthetic; click chemistry; photoaffinity labeling.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Insights Into Receptor-Based Anesthetic Pharmacophores and Anesthetic-Protein Interactions.Methods Enzymol. 2018;602:77-95. doi: 10.1016/bs.mie.2018.01.004. Epub 2018 Mar 2. Methods Enzymol. 2018. PMID: 29588042
-
Multiple propofol-binding sites in a γ-aminobutyric acid type A receptor (GABAAR) identified using a photoreactive propofol analog.J Biol Chem. 2014 Oct 3;289(40):27456-68. doi: 10.1074/jbc.M114.581728. Epub 2014 Aug 1. J Biol Chem. 2014. PMID: 25086038 Free PMC article.
-
Three classes of propofol binding sites on GABAA receptors.J Biol Chem. 2024 Oct;300(10):107778. doi: 10.1016/j.jbc.2024.107778. Epub 2024 Sep 11. J Biol Chem. 2024. PMID: 39270821 Free PMC article.
-
Mapping General Anesthetic Sites in Heteromeric γ-Aminobutyric Acid Type A Receptors Reveals a Potential For Targeting Receptor Subtypes.Anesth Analg. 2016 Nov;123(5):1263-1273. doi: 10.1213/ANE.0000000000001368. Anesth Analg. 2016. PMID: 27167687 Free PMC article. Review.
-
Multiple Non-Equivalent Interfaces Mediate Direct Activation of GABAA Receptors by Propofol.Curr Neuropharmacol. 2016;14(7):772-80. doi: 10.2174/1570159x14666160202121319. Curr Neuropharmacol. 2016. PMID: 26830963 Free PMC article. Review.
Cited by
-
Common general anesthetic propofol impairs kinesin processivity.Proc Natl Acad Sci U S A. 2017 May 23;114(21):E4281-E4287. doi: 10.1073/pnas.1701482114. Epub 2017 May 8. Proc Natl Acad Sci U S A. 2017. PMID: 28484025 Free PMC article.
-
Substrate-Dependent Modulation of SIRT2 by a Fluorescent Probe, 1-Aminoanthracene.Biochemistry. 2020 Oct 13;59(40):3869-3878. doi: 10.1021/acs.biochem.0c00564. Epub 2020 Sep 29. Biochemistry. 2020. PMID: 32941003 Free PMC article.
-
Alchemical Free Energy Calculations on Membrane-Associated Proteins.J Chem Theory Comput. 2023 Nov 14;19(21):7437-7458. doi: 10.1021/acs.jctc.3c00365. Epub 2023 Oct 30. J Chem Theory Comput. 2023. PMID: 37902715 Free PMC article. Review.
-
Structure-function Studies of GABA (A) Receptors and Related computer-aided Studies.J Mol Neurosci. 2023 Oct;73(9-10):804-817. doi: 10.1007/s12031-023-02158-3. Epub 2023 Sep 26. J Mol Neurosci. 2023. PMID: 37750966 Review.
-
Mechanistic basis of propofol-induced disruption of kinesin processivity.Proc Natl Acad Sci U S A. 2021 Feb 2;118(5):e2023659118. doi: 10.1073/pnas.2023659118. Proc Natl Acad Sci U S A. 2021. PMID: 33495322 Free PMC article.
References
-
- Baumann S. W., Baur R., and Sigel E. (2001) Subunit arrangement of γ-aminobutyric acid type A receptors. J. Biol. Chem. 276, 36275–36280 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

