Molecular Mechanisms of Enzyme Activation by Monovalent Cations
- PMID: 27462078
- PMCID: PMC5076497
- DOI: 10.1074/jbc.R116.737833
Molecular Mechanisms of Enzyme Activation by Monovalent Cations
Abstract
Regulation of enzymes through metal ion complexation is widespread in biology and underscores a physiological need for stability and high catalytic activity that likely predated proteins in the RNA world. In addition to divalent metals such as Ca2+, Mg2+, and Zn2+, monovalent cations often function as efficient and selective promoters of catalysis. Advances in structural biology unravel a rich repertoire of molecular mechanisms for enzyme activation by Na+ and K+ Strategies range from short-range effects mediated by direct participation in substrate binding, to more distributed effects that propagate long-range to catalytic residues. This review addresses general considerations and examples.
Keywords: enzyme kinetics; evolution; potassium; pyruvate kinase; sodium; structure-function; thrombin.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
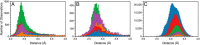
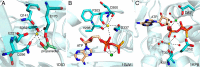
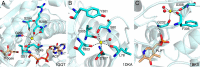
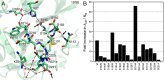
References
-
- Goswami T., Rolfs A., and Hediger M. A. (2002) Iron transport: emerging roles in health and disease. Biochem. Cell Biol. 80, 679–689 - PubMed
-
- Berridge M. J. (2012) Calcium signalling remodelling and disease. Biochem. Soc. Trans. 40, 297–309 - PubMed
-
- Di Cera E. (2006) A structural perspective on enzymes activated by monovalent cations. J. Biol. Chem. 281, 1305–1308 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

