AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity
- PMID: 27462105
- PMCID: PMC4987819
- DOI: 10.1073/pnas.1602419113
AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity
Abstract
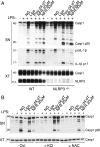
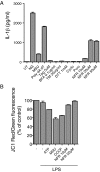
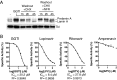
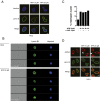
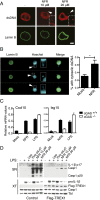
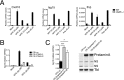
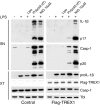
Inflammasomes are critical sensors that convey cellular stress and pathogen presence to the immune system by activating inflammatory caspases and cytokines such as IL-1β. The nature of endogenous stress signals that activate inflammasomes remains unclear. Here we show that an inhibitor of the HIV aspartyl protease, Nelfinavir, triggers inflammasome formation and elicits an IL-1R-dependent inflammation in mice. We found that Nelfinavir impaired the maturation of lamin A, a structural component of the nuclear envelope, thereby promoting the release of DNA in the cytosol. Moreover, deficiency of the cytosolic DNA-sensor AIM2 impaired Nelfinavir-mediated inflammasome activation. These findings identify a pharmacologic activator of inflammasome and demonstrate the role of AIM2 in detecting endogenous DNA release upon perturbation of nuclear envelope integrity.
Keywords: DNA sensors; Nelfinavir; inflammasome; nuclear envelope stress; zmpste24.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

