Sublethal Concentrations of Antibiotics Cause Shift to Anaerobic Metabolism in Listeria monocytogenes and Induce Phenotypes Linked to Antibiotic Tolerance
- PMID: 27462313
- PMCID: PMC4940397
- DOI: 10.3389/fmicb.2016.01091
Sublethal Concentrations of Antibiotics Cause Shift to Anaerobic Metabolism in Listeria monocytogenes and Induce Phenotypes Linked to Antibiotic Tolerance
Abstract
The human pathogenic bacterium Listeria monocytogenes is exposed to antibiotics both during clinical treatment and in its saprophytic lifestyle. As one of the keys to successful treatment is continued antibiotic sensitivity, the purpose of this study was to determine if exposure to sublethal antibiotic concentrations would affect the bacterial physiology and induce antibiotic tolerance. Transcriptomic analyses demonstrated that each of the four antibiotics tested caused an antibiotic-specific gene expression pattern related to mode-of-action of the particular antibiotic. All four antibiotics caused the same changes in expression of several metabolic genes indicating a shift from aerobic to anaerobic metabolism and higher ethanol production. A mutant in the bifunctional acetaldehyde-CoA/alcohol dehydrogenase encoded by lmo1634 did not have altered antibiotic tolerance. However, a mutant in lmo1179 (eutE) encoding an aldehyde oxidoreductase where rerouting caused increased ethanol production was tolerant to three of four antibiotics tested. This shift in metabolism could be a survival strategy in response to antibiotics to avoid generation of ROS production from respiration by oxidation of NADH through ethanol production. The monocin locus encoding a cryptic prophage was induced by co-trimoxazole and repressed by ampicillin and gentamicin, and this correlated with an observed antibiotic-dependent biofilm formation. A monocin mutant (ΔlmaDCBA) had increased biofilm formation when exposed to increasing concentration of co-trimoxazole similar to the wild type, but was more tolerant to killing by co-trimoxazole and ampicillin. Thus, sublethal concentrations of antibiotics caused metabolic and physiological changes indicating that the organism is preparing to withstand lethal antibiotic concentrations.
Keywords: Listeria monocytogenes; biofilm; gene expression; metabolism monocin; sublethal antibiotic concentrations.
Figures
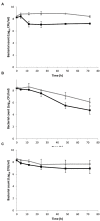
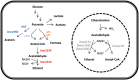
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

