G1/ELE Functions in the Development of Rice Lemmas in Addition to Determining Identities of Empty Glumes
- PMID: 27462334
- PMCID: PMC4941205
- DOI: 10.3389/fpls.2016.01006
G1/ELE Functions in the Development of Rice Lemmas in Addition to Determining Identities of Empty Glumes
Abstract
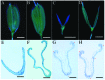
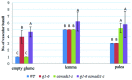
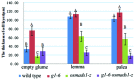
Rice empty glumes, also named sterile lemmas or rudimentary lemmas according to different interpretations, are distinct from lemmas in morphology and cellular pattern. Consistently, the molecular mechanism to control the development of lemmas is different from that of empty glumes. Rice LEAFY HULL STERILE1(OsLHS1) and DROOPING LEAF(DL) regulate the cellular pattern and the number of vascular bundles of lemmas respectively, while LONG STERILE LEMMA1 (G1)/ELONGATED EMPTY GLUME (ELE) and PANICLE PHYTOMER2 (PAP2)/OsMADS34 determine identities of empty glumes. Though some progress has been made, identities of empty glumes remain unclear, and genetic interactions between lemma genes and glume genes have been rarely elucidated. In this research, a new G1/ELE mutant g1-6 was identified and the phenotype was analyzed. Similar to previously reported mutant lines of G1/ELE, empty glumes of g1-6 plants transform into lemma-like organs. Furthermore, Phenotypes of single and double mutant plants suggest that, in addition to their previously described gene-specific functions, G1/ELE and OsLHS1 play redundant roles in controlling vascular bundle number, cell volume, and cell layer number of empty glumes and lemmas. Meanwhile, expression patterns of G1/ELE in osmads1-z flowers and OsLHS1 in g1-6 flowers indicate they do not regulate each other at the level of transcription. Finally, down-regulation of the empty glume gene OsMADS34/PAP2 and ectopic expression of the lemma gene DL, in the g1-6 plants provide further evidence that empty glumes are sterile lemmas. Generally, our findings provided valuable information for better understanding functions of G1 and OsLHS1 in flower development and identities of empty glumes.
Keywords: G1/ELE; OsLHS1; empty glume; lemma; rice.
Figures





Similar articles
-
ELE restrains empty glumes from developing into lemmas.J Genet Genomics. 2010 Feb;37(2):101-15. doi: 10.1016/S1673-8527(09)60029-1. J Genet Genomics. 2010. PMID: 20227044
-
The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the 'empty glumes' of rice (Oryza sativa) spikelets are in fact rudimentary lemmas.New Phytol. 2014 Apr;202(2):689-702. doi: 10.1111/nph.12657. Epub 2013 Dec 24. New Phytol. 2014. PMID: 24372518
-
The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet.Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):20103-8. doi: 10.1073/pnas.0907896106. Epub 2009 Nov 9. Proc Natl Acad Sci U S A. 2009. PMID: 19901325 Free PMC article.
-
Regulatory Role of OsMADS34 in the Determination of Glumes Fate, Grain Yield, and Quality in Rice.Front Plant Sci. 2016 Dec 15;7:1853. doi: 10.3389/fpls.2016.01853. eCollection 2016. Front Plant Sci. 2016. PMID: 28018389 Free PMC article.
-
Characterization and fine mapping of nonstop glumes 2 (nsg2) mutant in rice (Oryza sativa L.).Plant Biotechnol (Tokyo). 2019 Sep 25;36(3):125-134. doi: 10.5511/plantbiotechnology.19.0506a. Plant Biotechnol (Tokyo). 2019. PMID: 31768114 Free PMC article.
Cited by
-
Genome-Wide Identification and Characterization of wALOG Family Genes Involved in Branch Meristem Development of Branching Head Wheat.Genes (Basel). 2018 Oct 19;9(10):510. doi: 10.3390/genes9100510. Genes (Basel). 2018. PMID: 30347757 Free PMC article.
-
Characterization and Expression Analysis of the ALOG Gene Family in Rice (Oryza sativa L.).Plants (Basel). 2025 Apr 14;14(8):1208. doi: 10.3390/plants14081208. Plants (Basel). 2025. PMID: 40284096 Free PMC article.
-
Transcriptomics reveals a core transcriptional network of K-type cytoplasmic male sterility microspore abortion in wheat (Triticum aestivum L.).BMC Plant Biol. 2023 Dec 6;23(1):618. doi: 10.1186/s12870-023-04611-2. BMC Plant Biol. 2023. PMID: 38057735 Free PMC article.
-
Analysis of co-expression and gene regulatory networks associated with sterile lemma development in rice.BMC Plant Biol. 2023 Jan 6;23(1):11. doi: 10.1186/s12870-022-04012-x. BMC Plant Biol. 2023. PMID: 36604645 Free PMC article.
-
A hybrid sterile locus leads to the linkage drag of interspecific hybrid progenies.Plant Divers. 2020 Jul 25;42(5):370-375. doi: 10.1016/j.pld.2020.07.003. eCollection 2020 Oct. Plant Divers. 2020. PMID: 33134621 Free PMC article.
References
-
- Bell A. (1991). Plant Form: An Illustrated Guide to Flowering Plant Morphology. New York, NY: Oxford University Press.
-
- Chase A. (1968). The Structure of Grasses Explained for Beginners. Washington, DC: Smithsonian Institution.
LinkOut - more resources
Full Text Sources
Other Literature Sources

