Genome Wide Association Mapping in Arabidopsis thaliana Identifies Novel Genes Involved in Linking Allyl Glucosinolate to Altered Biomass and Defense
- PMID: 27462337
- PMCID: PMC4940622
- DOI: 10.3389/fpls.2016.01010
Genome Wide Association Mapping in Arabidopsis thaliana Identifies Novel Genes Involved in Linking Allyl Glucosinolate to Altered Biomass and Defense
Abstract
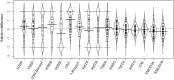
A key limitation in modern biology is the ability to rapidly identify genes underlying newly identified complex phenotypes. Genome wide association studies (GWAS) have become an increasingly important approach for dissecting natural variation by associating phenotypes with genotypes at a genome wide level. Recent work is showing that the Arabidopsis thaliana defense metabolite, allyl glucosinolate (GSL), may provide direct feedback regulation, linking defense metabolism outputs to the growth, and defense responses of the plant. However, there is still a need to identify genes that underlie this process. To start developing a deeper understanding of the mechanism(s) that modulate the ability of exogenous allyl GSL to alter growth and defense, we measured changes in plant biomass and defense metabolites in a collection of natural 96 A. thaliana accessions fed with 50 μM of allyl GSL. Exogenous allyl GSL was introduced exclusively to the roots and the compound transported to the leaf leading to a wide range of heritable effects upon plant biomass and endogenous GSL accumulation. Using natural variation we conducted GWAS to identify a number of new genes which potentially control allyl responses in various plant processes. This is one of the first instances in which this approach has been successfully utilized to begin dissecting a novel phenotype to the underlying molecular/polygenic basis.
Keywords: Arabidopsis; GWAS; allyl GSL; defense metabolism; novel genes; plant biomass.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

