A genome-wide IR-induced RAD51 foci RNAi screen identifies CDC73 involved in chromatin remodeling for DNA repair
- PMID: 27462432
- PMCID: PMC4860774
- DOI: 10.1038/celldisc.2015.34
A genome-wide IR-induced RAD51 foci RNAi screen identifies CDC73 involved in chromatin remodeling for DNA repair
Abstract
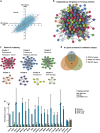
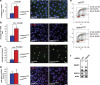
To identify new regulators of homologous recombination repair, we carried out a genome-wide short-interfering RNA screen combined with ionizing irradiation using RAD51 foci formation as readout. All candidates were confirmed by independent short-interfering RNAs and validated in secondary assays like recombination repair activity and RPA foci formation. Network analysis of the top modifiers identified gene clusters involved in recombination repair as well as components of the ribosome, the proteasome and the spliceosome, which are known to be required for effective DNA repair. We identified and characterized the RNA polymerase II-associated protein CDC73/Parafibromin as a new player in recombination repair and show that it is critical for genomic stability. CDC73 interacts with components of the SCF/Cullin and INO80/NuA4 chromatin-remodeling complexes to promote Histone ubiquitination. Our findings indicate that CDC73 is involved in local chromatin decondensation at sites of DNA damage to promote DNA repair. This function of CDC73 is related to but independent of its role in transcriptional elongation.
Keywords: CDC73; DNA repair; Parafibromin; RAD51; chromatin remodeling; genome-wide siRNA screen; homologous recombination; tumor suppressor.
Figures





References
-
- West SC . Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 2003; 4: 435–445. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

