Involvement of PARP1 in the regulation of alternative splicing
- PMID: 27462443
- PMCID: PMC4860959
- DOI: 10.1038/celldisc.2015.46
Involvement of PARP1 in the regulation of alternative splicing
Abstract
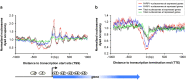
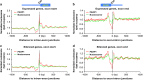
Specialized chromatin structures such as nucleosomes with specific histone modifications decorate exons in eukaryotic genomes, suggesting a functional connection between chromatin organization and the regulation of pre-mRNA splicing. Through profiling the functional location of Poly (ADP) ribose polymerase, we observed that it is associated with the nucleosomes at exon/intron boundaries of specific genes, suggestive of a role for this enzyme in alternative splicing. Poly (ADP) ribose polymerase has previously been implicated in the PARylation of splicing factors as well as regulation of the histone modification H3K4me3, a mark critical for co-transcriptional splicing. In light of these studies, we hypothesized that interaction of the chromatin-modifying factor, Poly (ADP) ribose polymerase with nucleosomal structures at exon-intron boundaries, might regulate pre-mRNA splicing. Using genome-wide approaches validated by gene-specific assays, we show that depletion of PARP1 or inhibition of its PARylation activity results in changes in alternative splicing of a specific subset of genes. Furthermore, we observed that PARP1 bound to RNA, splicing factors and chromatin, suggesting that Poly (ADP) ribose polymerase serves as a gene regulatory hub to facilitate co-transcriptional splicing. These studies add another function to the multi-functional protein, Poly (ADP) ribose polymerase, and provide a platform for further investigation of this protein's function in organizing chromatin during gene regulatory processes.
Keywords: Epigenetics; PARP1; chromatin; cotranscriptional splicing; gene regulation.
Figures






References
-
- Lopez AJ . Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet 1998; 32: 279–305. - PubMed
-
- Pan Q , Shai O , Lee LJ , Frey BJ , Blencowe BJ . Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40: 1413–1415. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

