Gene expression analysis of TIL rich HPV-driven head and neck tumors reveals a distinct B-cell signature when compared to HPV independent tumors
- PMID: 27462861
- PMCID: PMC5302866
- DOI: 10.18632/oncotarget.10788
Gene expression analysis of TIL rich HPV-driven head and neck tumors reveals a distinct B-cell signature when compared to HPV independent tumors
Abstract
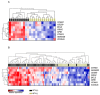
Human papilloma virus (HPV)-associated head and neck squamous cell carcinoma (HNSCC) has a better prognosis than it's HPV negative (HPV(-)) counterpart. This may be due to the higher numbers of tumor-infiltrating lymphocytes (TILs) in HPV positive (HPV(+)) tumors. RNA-Sequencing (RNA-Seq) was used to evaluate whether the differences in clinical behaviour simply reflect a numerical difference in TILs or whether there is a fundamental behavioural difference between TILs in these two settings. Thirty-nine HNSCC tumors were scored for TIL density by immunohistochemistry. After the removal of 16 TILlow tumors, RNA-Seq analysis was performed on 23 TILhigh/med tumors (HPV(+) n=10 and HPV(-) n=13). Using EdgeR, differentially expressed genes (DEG) were identified. Immune subset analysis was performed using Functional Analysis of Individual RNA-Seq/ Microarray Expression (FAIME) and immune gene RNA transcript count analysis. In total, 1,634 DEGs were identified, with a dominant immune signature observed in HPV(+) tumors. After normalizing the expression profiles to account for differences in B- and T-cell number, 437 significantly DEGs remained. A B-cell associated signature distinguished HPV(+) from HPV(-) tumors, and included the DEGs CD200, GGA2, ADAM28, STAG3, SPIB, VCAM1, BCL2 and ICOSLG; the immune signal relative to T-cells was qualitatively similar between TILs of both tumor cohorts. Our findings were validated and confirmed in two independent cohorts using TCGA data and tumor-infiltrating B-cells from additional HPV(+) HNSCC patients. A B-cell associated signal segregated tumors relative to HPV status. Our data suggests that the role of B-cells in the adaptive immune response to HPV(+) HNSCC requires re-assessment.
Keywords: RNA-sequencing; head and neck squamous cell carcinoma; human papilloma virus; transcriptome; tumor-infiltrating lymphocyte.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S, Bernstein L, Schoenberg JB, Stemhagen A, Fraumeni JF., Jr Smoking and drinking in relation to oral and pharyngeal cancer. Cancer research. 1988;48:3282–3287. - PubMed
-
- Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. Journal of clinical oncology. 2008;26:612–619. - PubMed
-
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. Journal of clinical oncology. 2011;29:4294–4301. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

