Tracking the Fragile X Mental Retardation Protein in a Highly Ordered Neuronal RiboNucleoParticles Population: A Link between Stalled Polyribosomes and RNA Granules
- PMID: 27462983
- PMCID: PMC4963131
- DOI: 10.1371/journal.pgen.1006192
Tracking the Fragile X Mental Retardation Protein in a Highly Ordered Neuronal RiboNucleoParticles Population: A Link between Stalled Polyribosomes and RNA Granules
Abstract
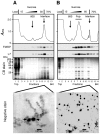
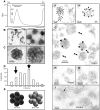
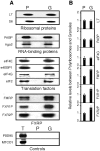
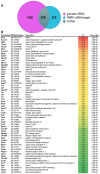
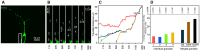
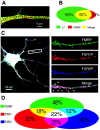
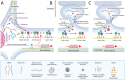
Local translation at the synapse plays key roles in neuron development and activity-dependent synaptic plasticity. mRNAs are translocated from the neuronal soma to the distant synapses as compacted ribonucleoparticles referred to as RNA granules. These contain many RNA-binding proteins, including the Fragile X Mental Retardation Protein (FMRP), the absence of which results in Fragile X Syndrome, the most common inherited form of intellectual disability and the leading genetic cause of autism. Using FMRP as a tracer, we purified a specific population of RNA granules from mouse brain homogenates. Protein composition analyses revealed a strong relationship between polyribosomes and RNA granules. However, the latter have distinct architectural and structural properties, since they are detected as close compact structures as observed by electron microscopy, and converging evidence point to the possibility that these structures emerge from stalled polyribosomes. Time-lapse video microscopy indicated that single granules merge to form cargoes that are transported from the soma to distal locations. Transcriptomic analyses showed that a subset of mRNAs involved in cytoskeleton remodelling and neural development is selectively enriched in RNA granules. One third of the putative mRNA targets described for FMRP appear to be transported in granules and FMRP is more abundant in granules than in polyribosomes. This observation supports a primary role for FMRP in granules biology. Our findings open new avenues for the study of RNA granule dysfunctions in animal models of nervous system disorders, such as Fragile X syndrome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Ramón y Cajal S. 1894. The Croonian lecture: la fine structure des centres nerveux. Proceedings of the Royal Society of London 55: 444–468.
-
- Abbott LF, Nelson SB. 2000. Synaptic plasticity: taming the beast. Nat Neurosci 3: 1178–1183. - PubMed
-
- Sutton MA, Schuman EM. 2006. Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127: 49–58. - PubMed
-
- Citri A, Malenka RC. 2008. Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacol 33: 18–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

