Rapamycin ameliorates chitosan nanoparticle-induced developmental defects of preimplantation embryos in mice
- PMID: 27463007
- PMCID: PMC5342693
- DOI: 10.18632/oncotarget.10813
Rapamycin ameliorates chitosan nanoparticle-induced developmental defects of preimplantation embryos in mice
Abstract
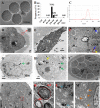
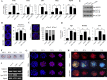
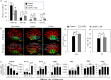
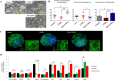
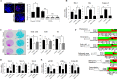
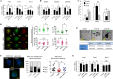
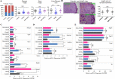
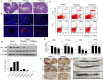
Chitosan nanoparticles (CSNPs) are used as drug or gene delivery vehicles. However, a detailed understanding of the effects of CSNPs on embryonic development remains obscure. Here, we show that CSNPs can be internalized into mouse blastocysts, such as the zona pellucida, the perivitelline space, and the cytoplasm. Consequently, CSNPs-induced endoplasmic reticulum (ER) stress increases both of Bip/Grp78, Chop, Atf4, Perk, and Ire1a mRNAs expression levels, and reactive oxygen species. Moreover, CSNPs show double- and multi-membraned autophagic vesicles, and lead to cell death of blastocoels. Conversely, treatment with rapamycin, which plays an important role as a central regulator of cellular proliferation and stress responses, decreased CSNPs-induced mitochondrial Ca+2 overloading, apoptosis, oxidative stress, ER stress, and autophagy. In vivo studies demonstrated that CSNPs injection has significant toxic effect on primordial and developing follicles. Notably, rapamycin rescued oxidative stress-induced embryonic defects via modulating gene expression of sirtuin and mammalian target of rapamycin. Interestingly, CSNPs treatment alters epigenetic reprogramming in mouse embryos. Overall, these observations suggest that rapamycin treatment could ameliorate CSNPs-induced developmental defects in preimplantation embryos. The data from this study would facilitate to understand the toxicity of these CSNPs, and enable the engineering of safer nanomaterials for therapeutic applications.
Keywords: ER stress; autophagy; chitosan nanoparticles; preimplantation; rapamycin.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures
Similar articles
-
The cytotoxic effects of dimethyl sulfoxide in mouse preimplantation embryos: a mechanistic study.Theranostics. 2017 Oct 17;7(19):4735-4752. doi: 10.7150/thno.21662. eCollection 2017. Theranostics. 2017. PMID: 29187900 Free PMC article.
-
α-Solanine induces ROS-mediated autophagy through activation of endoplasmic reticulum stress and inhibition of Akt/mTOR pathway.Cell Death Dis. 2015 Aug 27;6(8):e1860. doi: 10.1038/cddis.2015.219. Cell Death Dis. 2015. PMID: 26313911 Free PMC article.
-
Rapamycin attenuates visible light-induced injury in retinal photoreceptor cells via inhibiting endoplasmic reticulum stress.Brain Res. 2014 May 14;1563:1-12. doi: 10.1016/j.brainres.2014.02.020. Epub 2014 Mar 4. Brain Res. 2014. PMID: 24607296
-
Reactive oxygen species-mediated unfolded protein response pathways in preimplantation embryos.J Vet Sci. 2017 Mar 30;18(1):1-9. doi: 10.4142/jvs.2017.18.1.1. J Vet Sci. 2017. PMID: 28057903 Free PMC article. Review.
-
Molecular signal networks and regulating mechanisms of the unfolded protein response.J Zhejiang Univ Sci B. 2017 Jan.;18(1):1-14. doi: 10.1631/jzus.B1600043. J Zhejiang Univ Sci B. 2017. PMID: 28070992 Free PMC article. Review.
Cited by
-
Progeny Transfer Effects of Chitosan-Coated Cobalt Ferrite Nanoparticles.ACS Omega. 2023 Apr 19;8(17):15152-15159. doi: 10.1021/acsomega.3c00148. eCollection 2023 May 2. ACS Omega. 2023. PMID: 37151486 Free PMC article.
-
The cytotoxic effects of dimethyl sulfoxide in mouse preimplantation embryos: a mechanistic study.Theranostics. 2017 Oct 17;7(19):4735-4752. doi: 10.7150/thno.21662. eCollection 2017. Theranostics. 2017. PMID: 29187900 Free PMC article.
-
Zinc-Modified Nanotransporter of Doxorubicin for Targeted Prostate Cancer Delivery.Nanomaterials (Basel). 2017 Dec 8;7(12):435. doi: 10.3390/nano7120435. Nanomaterials (Basel). 2017. PMID: 29292780 Free PMC article.
-
Biodegradable and biocompatible high elastic chitosan scaffold is cell-friendly both in vitro and in vivo.Oncotarget. 2017 May 30;8(22):35583-35591. doi: 10.18632/oncotarget.14709. Oncotarget. 2017. PMID: 28103580 Free PMC article.
-
Reduced glutathione alleviates tunicamycin-induced endoplasmic reticulum stress in mouse preimplantation embryos.J Reprod Dev. 2018 Feb 27;64(1):15-24. doi: 10.1262/jrd.2017-055. Epub 2017 Oct 27. J Reprod Dev. 2018. PMID: 29081452 Free PMC article.
References
-
- Donaldson K. Resolving the nanoparticles paradox. Nanomedicine (Lond) 2006;1:229–234. - PubMed
-
- Kagan VE, Bayir H, Shvedova AA. Nanomedicine and nanotoxicology: two sides of the same coin. Nanomedicine. 2005;1:313–316. - PubMed
-
- Linkov I, Satterstrom FK, Corey LM. Nanotoxicology and nanomedicine: making hard decisions. Nanomedicine. 2008;4:167–171. - PubMed
-
- Kumar MNVR. A review of chitin and chitosan applications. React Funct Polym. 2000;46:1–27.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

