Comparative performance of transcriptome assembly methods for non-model organisms
- PMID: 27464550
- PMCID: PMC4964045
- DOI: 10.1186/s12864-016-2923-8
Comparative performance of transcriptome assembly methods for non-model organisms
Abstract
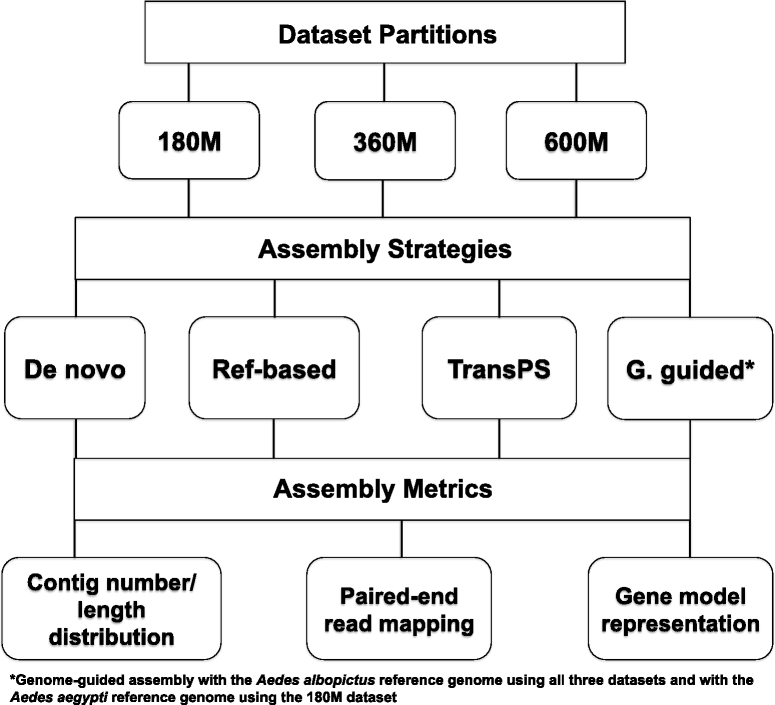
Background: The technological revolution in next-generation sequencing has brought unprecedented opportunities to study any organism of interest at the genomic or transcriptomic level. Transcriptome assembly is a crucial first step for studying the molecular basis of phenotypes of interest using RNA-Sequencing (RNA-Seq). However, the optimal strategy for assembling vast amounts of short RNA-Seq reads remains unresolved, especially for organisms without a sequenced genome. This study compared four transcriptome assembly methods, including a widely used de novo assembler (Trinity), two transcriptome re-assembly strategies utilizing proteomic and genomic resources from closely related species (reference-based re-assembly and TransPS) and a genome-guided assembler (Cufflinks).
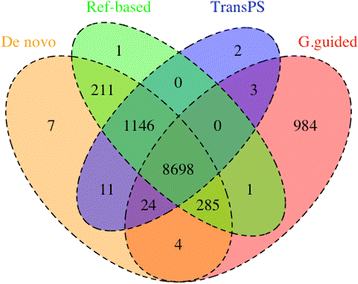
Results: These four assembly strategies were compared using a comprehensive transcriptomic database of Aedes albopictus, for which a genome sequence has recently been completed. The quality of the various assemblies was assessed by the number of contigs generated, contig length distribution, percent paired-end read mapping, and gene model representation via BLASTX. Our results reveal that de novo assembly generates a similar number of gene models relative to genome-guided assembly with a fragmented reference, but produces the highest level of redundancy and requires the most computational power. Using a closely related reference genome to guide transcriptome assembly can generate biased contig sequences. Increasing the number of reads used in the transcriptome assembly tends to increase the redundancy within the assembly and decrease both median contig length and percent identity between contigs and reference protein sequences.
Conclusions: This study provides general guidance for transcriptome assembly of RNA-Seq data from organisms with or without a sequenced genome. The optimal transcriptome assembly strategy will depend upon the subsequent downstream analyses. However, our results emphasize the efficacy of de novo assembly, which can be as effective as genome-guided assembly when the reference genome assembly is fragmented. If a genome assembly and sufficient computational resources are available, it can be beneficial to combine de novo and genome-guided assemblies. Caution should be taken when using a closely related reference genome to guide transcriptome assembly. The quantity of read pairs used in the transcriptome assembly does not necessarily correlate with the quality of the assembly.
Keywords: Aedes albopictus; De novo assembly; Genome-guided assembly; Next-generation sequencing; Non-model organisms; Reference-based re-assembly; TransPS; Transcriptome assembly.
Figures


References
-
- Genome 10K Project. https://genome10k.soe.ucsc.edu/. Accessed 7 Apr 2015.
-
- i5k Genome Sequencing Initiative for Insects and Other Arthropods. http://www.arthropodgenomes.org/wiki/i5K. Accessed 7 Apr 2015.
-
- Yang HJ, Ratnapriya R, Cogliati T, Kim JW, Swaroop A. Vision from next generation sequencing: Multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog Retin Eye Res. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

