flowVS: channel-specific variance stabilization in flow cytometry
- PMID: 27465477
- PMCID: PMC4964071
- DOI: 10.1186/s12859-016-1083-9
flowVS: channel-specific variance stabilization in flow cytometry
Abstract
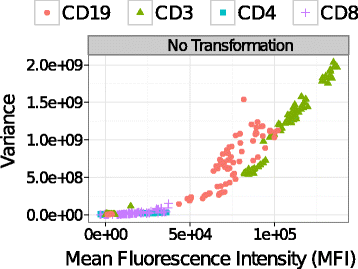
Background: Comparing phenotypes of heterogeneous cell populations from multiple biological conditions is at the heart of scientific discovery based on flow cytometry (FC). When the biological signal is measured by the average expression of a biomarker, standard statistical methods require that variance be approximately stabilized in populations to be compared. Since the mean and variance of a cell population are often correlated in fluorescence-based FC measurements, a preprocessing step is needed to stabilize the within-population variances.
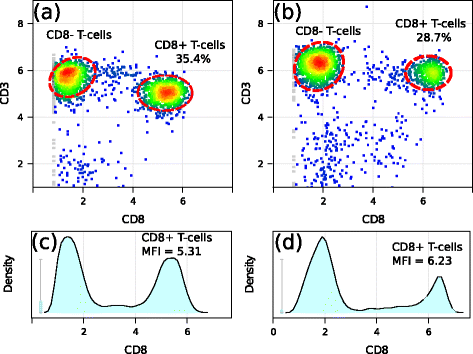
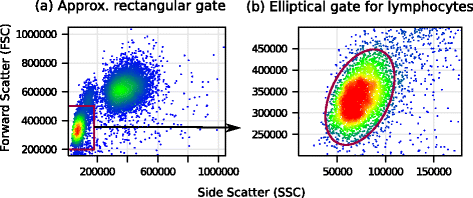
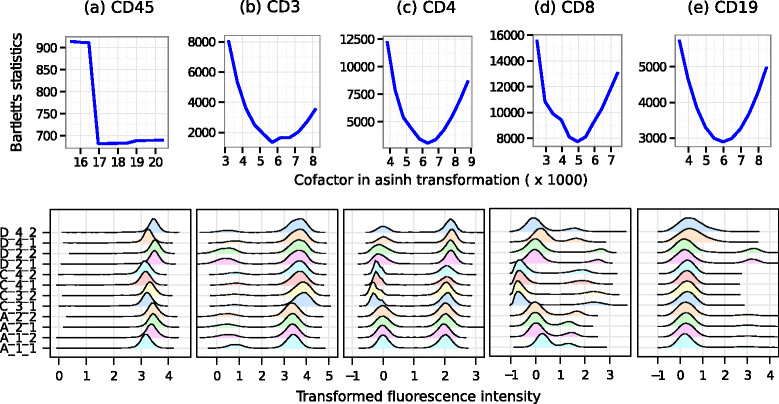
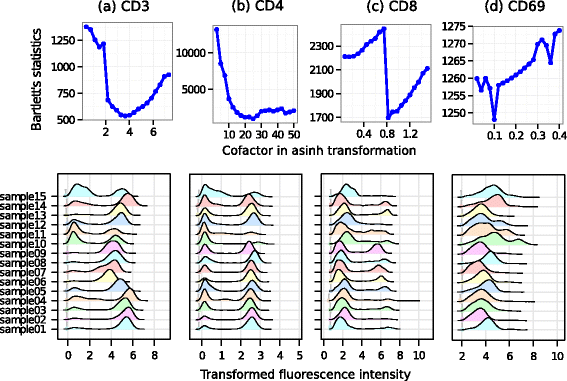
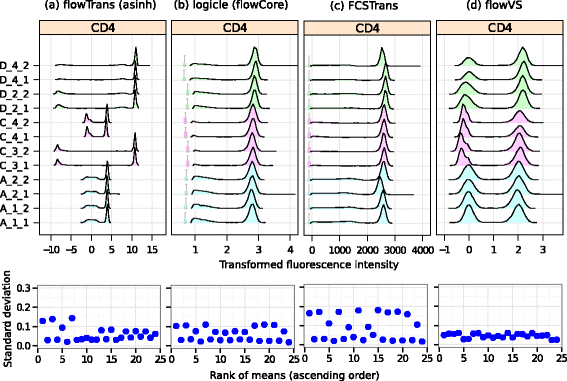
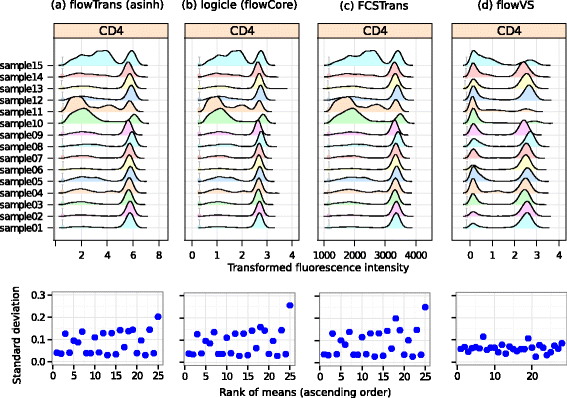
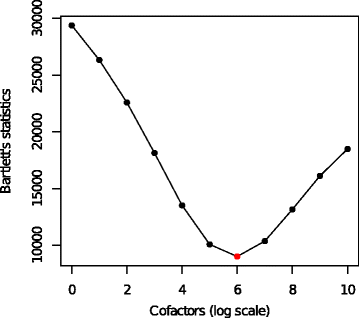
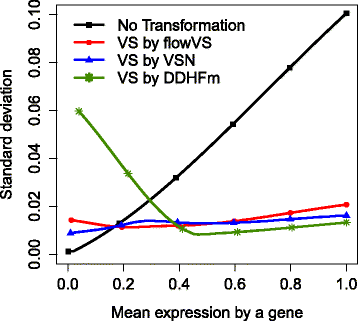
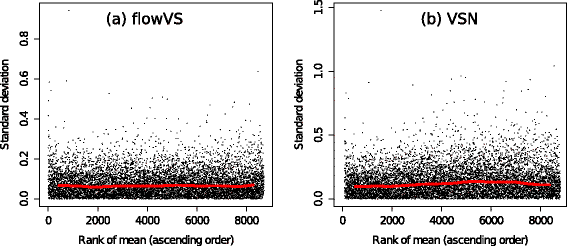
Results: We present a variance-stabilization algorithm, called flowVS, that removes the mean-variance correlations from cell populations identified in each fluorescence channel. flowVS transforms each channel from all samples of a data set by the inverse hyperbolic sine (asinh) transformation. For each channel, the parameters of the transformation are optimally selected by Bartlett's likelihood-ratio test so that the populations attain homogeneous variances. The optimum parameters are then used to transform the corresponding channels in every sample. flowVS is therefore an explicit variance-stabilization method that stabilizes within-population variances in each channel by evaluating the homoskedasticity of clusters with a likelihood-ratio test. With two publicly available datasets, we show that flowVS removes the mean-variance dependence from raw FC data and makes the within-population variance relatively homogeneous. We demonstrate that alternative transformation techniques such as flowTrans, flowScape, logicle, and FCSTrans might not stabilize variance. Besides flow cytometry, flowVS can also be applied to stabilize variance in microarray data. With a publicly available data set we demonstrate that flowVS performs as well as the VSN software, a state-of-the-art approach developed for microarrays.
Conclusions: The homogeneity of variance in cell populations across FC samples is desirable when extracting features uniformly and comparing cell populations with different levels of marker expressions. The newly developed flowVS algorithm solves the variance-stabilization problem in FC and microarrays by optimally transforming data with the help of Bartlett's likelihood-ratio test. On two publicly available FC datasets, flowVS stabilizes within-population variances more evenly than the available transformation and normalization techniques. flowVS-based variance stabilization can help in performing comparison and alignment of phenotypically identical cell populations across different samples. flowVS and the datasets used in this paper are publicly available in Bioconductor.
Keywords: Bartlett’s test; Flow cytometry; Microarrays; Variance stabilization.
Figures

References
-
- Peters JM, Ansari MQ. Multiparameter flow cytometry in the diagnosis and management of acute leukemia. Arch Pathol Lab Med. 2011;135(1):44–54. - PubMed
-
- Azad A, Rajwa B, Pothen A. Immunophenotypes of acute myeloid leukemia from flow cytometry data using templates. 2014. http://arxiv.org/abs/1403.6358.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

