IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells
- PMID: 27465917
- PMCID: PMC5025899
- DOI: 10.1182/blood-2016-02-698027
IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells
Abstract
Treatment of hematological malignancies by adoptive transfer of activated natural killer (NK) cells is limited by poor postinfusion persistence. We compared the ability of interleukin-2 (IL-2) and IL-15 to sustain human NK-cell functions following cytokine withdrawal to model postinfusion performance. In contrast to IL-2, IL-15 mediated stronger signaling through the IL-2/15 receptor complex and provided cell function advantages. Genome-wide analysis of cytosolic and polysome-associated messenger RNA (mRNA) revealed not only cytokine-dependent differential mRNA levels and translation during cytokine activation but also that most gene expression differences were primed by IL-15 and only manifested after cytokine withdrawal. IL-15 augmented mammalian target of rapamycin (mTOR) signaling, which correlated with increased expression of genes related to cell metabolism and respiration. Consistently, mTOR inhibition abrogated IL-15-induced cell function advantages. Moreover, mTOR-independent STAT-5 signaling contributed to improved NK-cell function during cytokine activation but not following cytokine withdrawal. The superior performance of IL-15-stimulated NK cells was also observed using a clinically applicable protocol for NK-cell expansion in vitro and in vivo. Finally, expression of IL-15 correlated with cytolytic immune functions in patients with B-cell lymphoma and favorable clinical outcome. These findings highlight the importance of mTOR-regulated metabolic processes for immune cell functions and argue for implementation of IL-15 in adoptive NK-cell cancer therapy.
© 2016 by The American Society of Hematology.
Figures
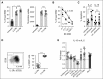


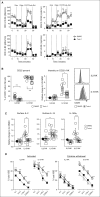
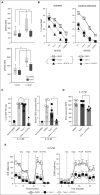
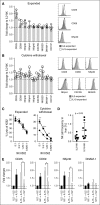

Similar articles
-
Deciphering Natural Killer Cell Homeostasis.Front Immunol. 2020 May 12;11:812. doi: 10.3389/fimmu.2020.00812. eCollection 2020. Front Immunol. 2020. PMID: 32477340 Free PMC article. Review.
-
Autonomous growth and increased cytotoxicity of natural killer cells expressing membrane-bound interleukin-15.Blood. 2014 Aug 14;124(7):1081-8. doi: 10.1182/blood-2014-02-556837. Epub 2014 Jul 8. Blood. 2014. PMID: 25006133
-
TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway.Sci Signal. 2016 Feb 16;9(415):ra19. doi: 10.1126/scisignal.aad1884. Sci Signal. 2016. PMID: 26884601
-
Preactivation with IL-12, IL-15, and IL-18 induces CD25 and a functional high-affinity IL-2 receptor on human cytokine-induced memory-like natural killer cells.Biol Blood Marrow Transplant. 2014 Apr;20(4):463-73. doi: 10.1016/j.bbmt.2014.01.006. Epub 2014 Jan 13. Biol Blood Marrow Transplant. 2014. PMID: 24434782 Free PMC article.
-
The regulation and biological activity of interleukin 12.Leuk Lymphoma. 1998 May;29(5-6):427-38. doi: 10.3109/10428199809050903. Leuk Lymphoma. 1998. PMID: 9643557 Review.
Cited by
-
Modulating HIV-1 envelope glycoprotein conformation to decrease the HIV-1 reservoir.Cell Host Microbe. 2021 Jun 9;29(6):904-916.e6. doi: 10.1016/j.chom.2021.04.014. Epub 2021 May 20. Cell Host Microbe. 2021. PMID: 34019804 Free PMC article.
-
Deciphering Natural Killer Cell Homeostasis.Front Immunol. 2020 May 12;11:812. doi: 10.3389/fimmu.2020.00812. eCollection 2020. Front Immunol. 2020. PMID: 32477340 Free PMC article. Review.
-
Immunomodulatory Effects of IL-2 and IL-15; Implications for Cancer Immunotherapy.Cancers (Basel). 2020 Nov 30;12(12):3586. doi: 10.3390/cancers12123586. Cancers (Basel). 2020. PMID: 33266177 Free PMC article. Review.
-
Transcription Factors Associated With IL-15 Cytokine Signaling During NK Cell Development.Front Immunol. 2021 Mar 18;12:610789. doi: 10.3389/fimmu.2021.610789. eCollection 2021. Front Immunol. 2021. PMID: 33815365 Free PMC article. Review.
-
Strategies to Augment Natural Killer (NK) Cell Activity against Solid Tumors.Cancers (Basel). 2019 Jul 23;11(7):1040. doi: 10.3390/cancers11071040. Cancers (Basel). 2019. PMID: 31340613 Free PMC article. Review.
References
-
- Miller JS, Soignier Y, Panoskaltsis-Mortari A, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105(8):3051–3057. - PubMed
-
- Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097–2100. - PubMed
-
- Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97(1):14–32. - PubMed
-
- Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6(8):595–601. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

